Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Advances in light microscopy techniques and labelling approaches have allowed for much greater flexibility in terms of experimental capabilities within virology research. Super resolution techniques such as STORM, PALM, SIM, TIRF and SRRF-Stream+ allow the diffraction limit of light to be overcome and improve imaging resolution down to 50 nm, or beyond. Improved resolution affords greater insight into the intricate and dynamic nature of how viruses interact with the host cell during the infection cycle. The range of what can be studied using light microscopy techniques can therefore span from studying the general histopathological effects within tissue samples, detection of virus within a sample, localization of viruses within cells and potentially, tracking of live viruses during the virus infection cycle.
A fundamental aspect of handling any potential samples containing a known virus within the laboratory, or indeed an unknown virus or pathogen in a clinical setting, is of course safety. Samples should be prepared for analysis according to the biosafety guidelines for the virus under study. It should be noted that this can vary from one region to another since a specific virus may be considered endemic within a population or present a higher risk in another. The higher the biosafety level of the laboratory the greater the restrictions there is to the equipment and indeed what experiments can be performed. Often this will mean that the virus needs to be inactivated (e.g. using heat, chemical or pH treatments) before it can be used in the laboratory in labelling and imaging experiments. As an alternative, it may be possible to use a closely related “low-risk” virus, use virus-like particles (VLPs), or other strategies as a research model.
ELISA and PCR methods can be used to detect viruses in a sample and make the basis of many diagnostic detection and rapid screening techniques for HIV, influenza or SARS -CoV2. However, these techniques do not let us visualize viruses or their interactions with host-cells. Thus, it is not possible to understand much about the infection process itself. For this, Immunostaining or Immunofluorescence labelling can be used so that proteins of interest, of both the cell and virus, can be labelled with fluorescently tagged antibodies. Single molecule FISH (Fluorescence in situ Hybridization) methods may also be used to probe, and tag short sequences of the viral genetic material.
Firstly, the sample needs to be fixed to maintain the structure of the cell or tissue being studied. This involves agents such as acetone, methanol, formaldehyde, or paraformaldehyde to fix the cells and virus. Then a suitable antibody with a fluorescent label added, with unbound antibody removed through a series of washes. The cells may then be observed using a suitable light microscopy technique.
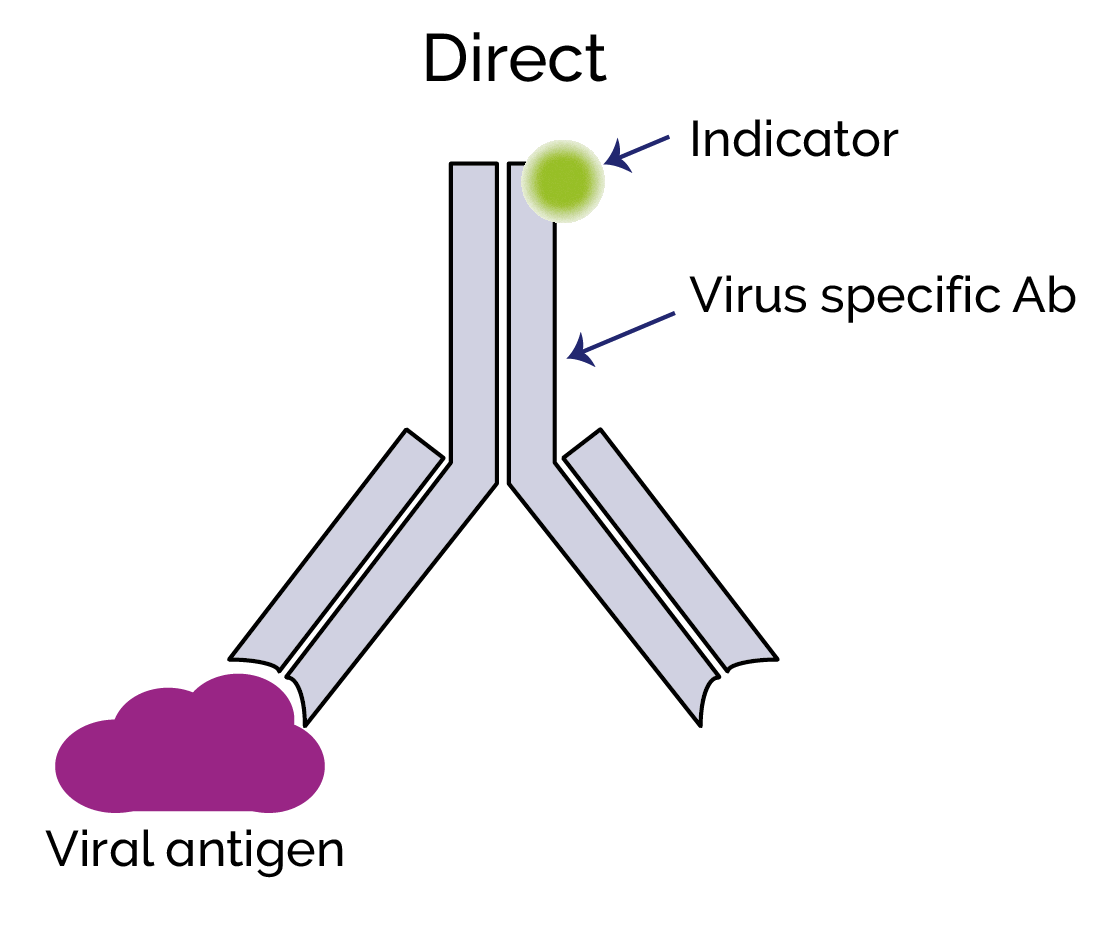
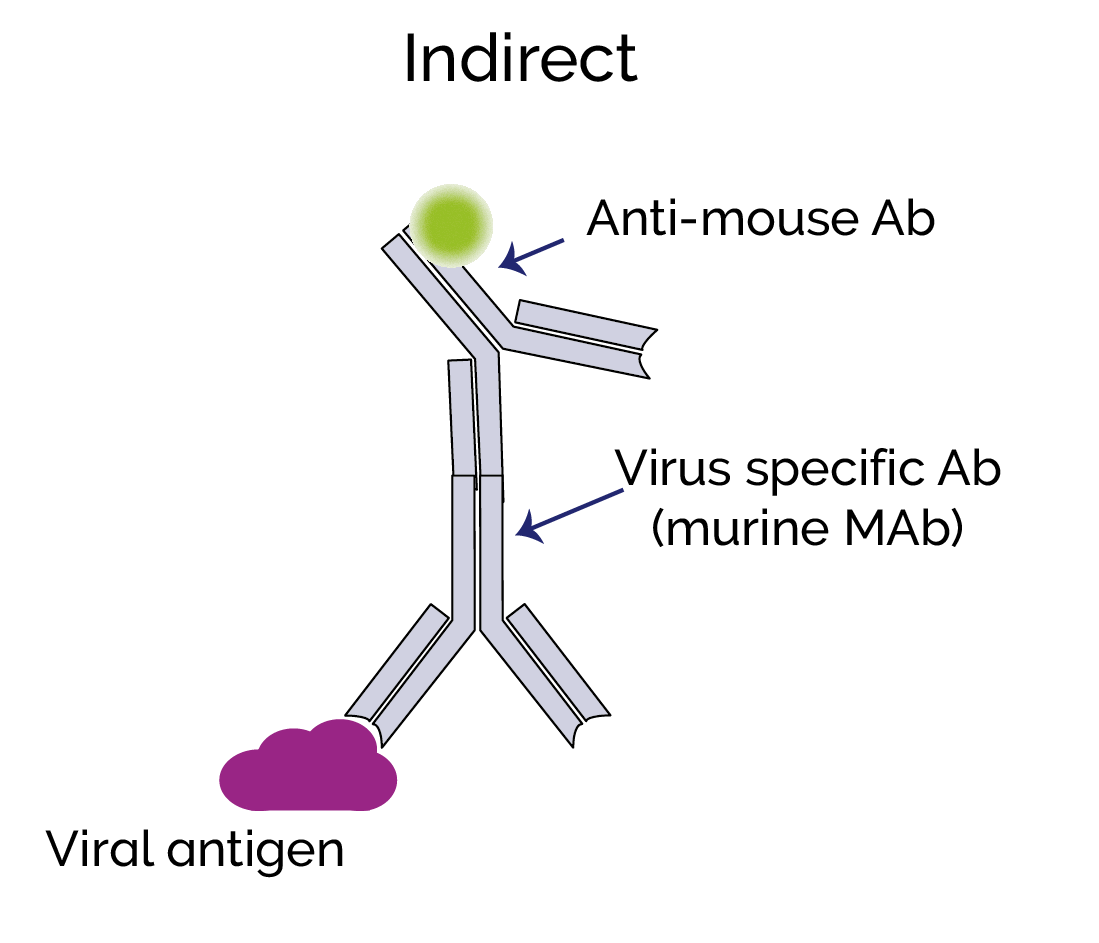
Direct immunolabelling involves the direct binding of a fluorescent label to a viral antigen. Indirect immunolabelling uses a secondary antibody that will bind to the virus-specific primary antibody. Since many secondary antibodies with the fluorescent label can potentially bind to the primary virus-specific antibody, this can produce a much higher signal level than direct labelling.
Live-cell imaging is more challenging than imaging of fixed cells; with fixed cells, you may be able to use longer exposures and capture a stronger signal. However, it is still possible to do live-imaging of viruses and we can now choose from a number of genetically encoded fluorescent proteins to label specific viral components such as envelope proteins or nucleocapsid. Immunolabelling is generally not suitable for live cell imaging, but any labelling can interfere with the viability of the virus and normal function of the virus components. This must be considered and validated in the experimental design. A fluorescent protein can be fused to a target protein by inserting the code for the label into the open reading frame (ORF) of the target protein, for example, a virus nucleocapsid protein. Ideally this should be so there are enough copies per virion so that a useful signal level can be detected using high sensitivity EMCCD or sCMOS cameras. The host-cell can also be labelled, and then virus-host-cell interactions can thus be studied e.g. cell membrane, cytoplasm or nucleus.
Viruses are very small relative to the size of the cells they infect. Medium sized viruses like Adenovirus, Coronavirus or Influenza virus are in the order of 90-125 nm in size. Small viruses such as Parvovirus can be ~20 nm, and others as small 5 nm in size. This means that only a small number of labels can bind to the virus before self-quenching effects happen to the closely bound labels. As highlighted in the previous paragraphs, incorporation and expression of fluorescent proteins may also impact normal function of the virus due to steric hinderance, and the interactions with the cell.
Commonly used Fluorescent labels include:
Fluorescent dyes which are small, organic natural or synthetic fluorescent molecules e.g. fluorescein, DAPI, Texas red, TRITC, Cy5 and Alexa fluor
Fluorescent Proteins – are larger proteins that fluoresce due to their structure. e.g. GFP (Green Fluorescent protein).
In considering labels, options that emit more towards the red spectrum, would mean that autofluorescence from the host-cell making it easier to obtain a better signal to noise from the low virus label signals against the background. Therefore, Cy5, mCherry or some of the Alexa Fluor labels with longer emission wavelengths could make good candidates for labelling viruses. It should be noted that optics of older microscopes may not be suitable wavelengths into the Near Infrared spectrum.
Quantum Dots (Qdots) are semiconductor crystals that are 2-10 nm in size. Qdots provide a brighter signal than conventional fluorescent labels, are stable and have discrete emission spectra. They have been widely used for single molecule studies and proven very useful due to their optical attributes. This also makes them very interesting for potential application in virology studies. To date despite this, there has been a slow uptake of Qdots for virology studies, in part due to difficulties with labelling the viruses effectively and without perturbation of normal infectivity. However, this is changing and Qdots are starting to be used by a growing number of research groups.
The following table lists some of the main light microscopy techniques compared with important experimental parameters. This can be considered as a general guide to the characteristics of different techniques. In some cases variations of the original techniques listed may have been developed to improve aspects such as speed of imaging, or the ability to image in 3D, or live cell applications.
| Parameter | Widefield Epi-fluorescence | Confocal Laser Scanning Microscopy |
Confocal Spinning Disk Microscopy* | Dragonfly Multimodal System | TIRF | Multi-photon Microscopy | Light-Sheet (SPIM) | SIM | STED | PALM/STORM | SRRF-Stream+ |
| Live Imaging Experiments | ■ | ■ | ■ | ■ | ■ | ■ | □ | ■ | ■ | ■ | |
| Imaging over long durations | ■ | □ | ■ | ■ | ■ | ■ | |||||
| High Speed Imaging | ■ | □ | ■ | ■ | |||||||
| Super-resolution | ■ | ■ | ■ | ■ | ■ | ■ | ■ | ||||
| Suitable for FRET Studies | ■ | ■ | ■ | ■ | ■ | ||||||
| Image in 3D | □ | ■ | ■ | ■ | ■ | ■ | ■ | □ | |||
| Image deep Tissues | ■ | ■ | ■ | ||||||||
| Comments | Limited resolution. High speeds and simple. | Improved SNR over widefield for greater image clarity. | Dragonfly imaging system combines multiple techniques. | Improved resolution in Z axis and can be combined with other techniques | Useful for relatively thick samples. | Useful for relatively thick samples. Limited resolution. | Moderate resolution improvement, using normal fluorophores | High resolution over small field of view. Complex optical set-up. | High resolution – but requires specific labels. | Live cell imaging at low illumination intensities with any fluorophores | |
Different light microscopy techniques used across 100 recent virology publications with the iXon EMCCD camera (2017-2020) alongside new Dragonfly System and SRRF-Stream+. The applicability of each technique is compared against different experimental parameters. ■ suitable, some variants may be suitable.
Widefield fluorescence or more accurately epifluorescence microscopy is the most simple and accessible technique since it may be performed using a standard microscope and sensitive camera, allowing live cell imaging at high speeds and over a depth of field and wide field of view. However, it is limited to classical resolution limits meaning that it will not be suitable when higher resolution is required. In addition, the out of focus background fluorescence will reduce the signal to noise and so it will only be possible to detect very strong label signals.
Confocal Microscopy will further improve the signal to noise for much improved image clarity by eliminating out of focus light and potentially allowing live and 3D imaging. Confocal also will require a high sensitivity detector for the low signal levels inherent to this technique and of the viruses themselves. There are two main types of confocal Microscopy – laser scanning and Spinning disk. Find out more in the confocal microscopy overview article.
TIRF (Total Internal Reflectance) microscopy is a common technique used in virology research since it enables enhanced resolution with a high signal to noise. The downside of this technique is that the depth of sample is limited to ~100nm depth of the evanescent wave. This means that TIRF is typically used for imaging the stages of the virus cycle close to the cell membrane i.e. attachment, membrane fusion and egress of viral progeny from the host cell. Find out more about TIRF in the TIRF Microscopy article.
Stochastic Optical Reconstruction Microscopy (STORM), photoactivated localization microscopy (PALM) and Stimulated emission depletion (STED) offer exceptional resolution abilities. This makes it possible to image labelled viruses in a way that was not possible before. Variants of STORM, PALM and STED have continued to be developed so that they can allow live and/or 3D imaging. There are understandably sacrifices that must be made to achieve these resolutions.
STORM and PALM use blinking fluorophores to localize the point-source emissions from the rapidly switching fluorophores from their dark state with high precision allowing a high-resolution image to be reconstructed from 100 s to 1000 s of frames. With these techniques resolutions of 20-50 nm are possible. The use of specific photo-switchable fluorophores is a limitation to these techniques since it restricts what labels can be used.
In STED, a high-intensity laser excites the sample and stimulates fluorescent emission. A second laser acts in a “donut” shape to deplete fluorescence except for that occurring in a small central sub-region of ~30-80 nm. The instrument scans across the sample building up a super-resolved image.
A downside of this technique is the complex optical set-up, and that high laser powers are required potentially causing phototoxicity to the cells. However, STED can produce relatively fast, high-resolution images over a small field of view which can be suitable for some virology experiments.
SIM – Structured Illumination Microscopy is another imaging technique that may be used for imaging viruses and host-cells. Unlike some other super-resolution methods e.g. STORM or PALM, this method uses an optical means to achieve enhanced resolution. Illumination is applied in a sequence of rotating spatially spaced patterns to create optical interference, and the Moire patterns generated can be processed (via Fourier-based analysis) to create an image with improved resolution. Further variations add a further dimension and this allow for 3D-SIM. The resolution of SIM of 120-150 nm is not as good as STORM, PALM or STED. However, since SIM is a purely optical approach you are not restricted to specific labels and the technique can prove to be a flexible way to achieve double the resolution of epi-fluorescence microscopy.
SRRF (Super Resolution Radial Fluctuations) is a technique that uses the natural fluctuations in fluorescence over time to position the emission according to their radiality. SRRF-Stream+ is a further development of this technique that allows the process to be run in real-time. 100 short exposure frames will create a super-resolved image. This technique can work on multiple imaging modalities – widefield, confocal or TIRF to enhance resolution to between 50 to 150 nm depending on the sample, settings and microscope. It allows live imaging of cells using low illumination intensities and is highly flexible making this technique potentially suitable to many experiments using fixed or live cells. Find out more about SRRF-Stream+.
The Andor Dragonfly system is a unique imaging solution since it allows multiple imaging modalities to be selected depending on the specific experiment. This means it is possible to switch between rapid and convenient wide-field imaging, or use increase resolution using confocal, TIRF, dSTORM or SRRF-Stream+. Find out more about Dragonfly.
FRET (Fluorescence Resonance Energy Transfer) enables the proximity of two fluorophores to be determined within nanometre precision. FRET can be used with confocal and TIRF imaging modalities so that it is possible to get information on spatial and temporal relationships in living cells. FRET is therefore very useful for investigating and tracking many of the dynamic interactions of virions throughout the infection cycle. There are many examples of use including: showing viral RNA and proteins to be co-localised with components of the host cell membrane, cytoskeleton or nucleus, study of replication events, assembly of virions and uncoating of capsids. Find out more about FRET.
A key part of microscopy techniques are the imaging detectors. Ever more sensitive detector technologies have helped make these, and other light microscopy techniques, applicable to virology studies which typically have very weak signal levels. EMCCD detectors such as the Andor iXon cameras have been widely used. These cameras can detect single photons and thus, are perfectly suited to the demands of confocal, TIRF and STORM. In recent years sCMOS camera technology, the most common detector technology for general cell biology studies has improved further and are more relevant to a wider range of virology imaging experiments. This has been further aided by new, brighter fluorophores and improved labelling strategies. The latest generation of back-illuminated sCMOS cameras such as the Sona series offer an alternative option to EMCCD cameras, providing higher temporal resolution over wider fields of views. For where the weakest signals are involved, EMCCD cameras like the latest iXon Ultra and Life EMCCD cameras remain the most suitable detector option as the sensitivity remains unmatched via other competing technologies.
Maolin Lu, Xiaochu Ma, Walther Mothes, Chapter Eight - Illuminating the virus life cycle with single-molecule FRET imaging, Editor(s): Félix A. Rey, Advances in Virus Research, Academic Press, Volume 105, 2019, Pages 239-273, ISSN 0065-3527, ISBN 9780128184561,
