Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Whole Cell 3D Super-Resolution Imaging by ‘STORM’
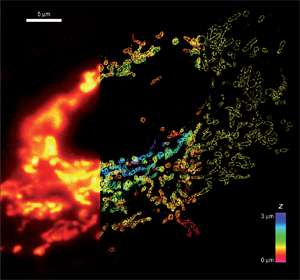
{Figure 1: Comparison of conventional and STORM images of mitochondria in a mammalian cell. The mitochondrial outer membrane protein Tom20 was labeled. (left panel) Conventional image of the left part of the cell. (middle panel) 3D STORM image of the middle part of the cell. The z-dimension information is color-coded according to the color scale bar. (right panel). The xy cross-section of the STORM image of the right part of the cell. Image courtesy of Zhuang Research Group, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA.}
Resolution limits imposed by classical light diffraction represent a major limitation in the ability to obtain meaningful insights into the fabric of living cells. Conventional optical microscopy tools have only revealed structures measuring approximately 200 nm across. Unfortunately, most cellular organelles involved in physiological processes including cell-to-cell communication, cell growth and division are often below this limiting threshold. In fact, many cytoskeletal assemblies can be smaller than 50 nm and our understanding of their function could be greatly improved if direct optical access to that nano realm was readily available.
A novel 3D super-resolution microscopic technique has made it possible to gain new insights into previously unresolvable environments. The approach of 3D Stochastic Optical Reconstruction Microscopy (‘STORM’) described by the Zhuang lab, has recently enabled the team to image whole cells in multi-color. Using multiple photoswitchable tags, the entire mitochondrial network of fixed mammalian cells has been interrogated, elucidating mitochondrial morphology and microtubule interactions which were obscured in conventional fluorescence images.
The STORM approach uses sequential imaging of single fluorophore molecules as they toggle between bright and dark states. By exciting only a stochastic subset of single labels with an activating pulse of laser, one obtains a low light image of individual molecules that can be discerned as single diffraction-limited spots. This allows the position of each fluorescent molecule to be determined with nanometer precision. Such repeated cycles of pulses allow the position of all molecules to be determined, and subsequently the construction of a super-resolution image from these precisely determined fluorophore positions.
Two-dimension images with 20-30 nm lateral resolution were initially obtained. Extension to 3D localization has been achieved by introducing an ‘astigmatic’ imaging method which generates two slightly different focal planes, allowing axial positions of the molecules to be determined to an accuracy of 50-60 nm by analysing the ellipticity and orientation of the fluorophore image.
However, nanoscopic recordings involve imaging a limited number of isolated molecules per frame. Furthermore, short exposure times and fast frame rates are essential to acquire the necessary composite image set within a time-frame that will ultimately make the technique usable for following dynamic processes in living cells.
Recent advancements in super-resolution have been possible with Electron Multiplying Charge-Coupled Device (EMCCD) technology from Andor. In low light conditions image acquisition is a challenging task due to low-signal/high-noise nature of such experiments and the prime advantage of Andor′s high-performance iXon Ultra 888 EMCCD camera is it's ability to minimize the camera noise floor, even at fast readout speeds. Special features such as ‘optically centred cropped sensor mode’ acquisition, combined with optimal readout electronics, ensure that iXon Ultra 888 can also deliver industry leading frame rates.
