Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Plasma membranes are supramolecular structures that play important roles in cellular processes and are the border between the cellular exterior and interior over which all transport from information over nutrients to infectious agents takes place. Plasma membrane dynamics and organization are determining factors in supporting and regulating these processes.
Typically, studies of plasma membrane, and other processes of the cell may focus on the structural and organization aspects, or the dynamics involved, but seldom both at the same time. It is only by having both aspects of a biological process that we cab really really gain a complete understanding- so how can this be done?
Professor Thorsten Wohland is group leader of the Biophysical Fluorescence Lab (BFL) at the Department of Biological Sciences & Chemistry at the National university of Singapore (NUS). Their lab is interested in precisely these kinds of questions, developing and sharing, fluorescence, single molecule sensitive imaging and spectroscopy techniques for study of complex cellular processes.
“Structure and Dynamics are two sides of the same coin. Only by recording both can we obtain a full understanding of a biological system.” - Professor Thorsten Wohland
In the Biophysical Fluorescence Lab at NUS, we have developed imaging fluorescence correlation spectroscopy (ImFCS) to study these cellular membranes. ImFCS is a camera-based modality that allows quantitative spatiotemporal analysis of molecular dynamics over thousands of contiguous points providing spatiotemporal information to determine membrane dynamics and organization. It uses fast and sensitive EMCCD and sCMOS cameras and open-source software for data acquisition and evaluation. A workflow of a typical imaging FCS experiment is shown in Figure 1.
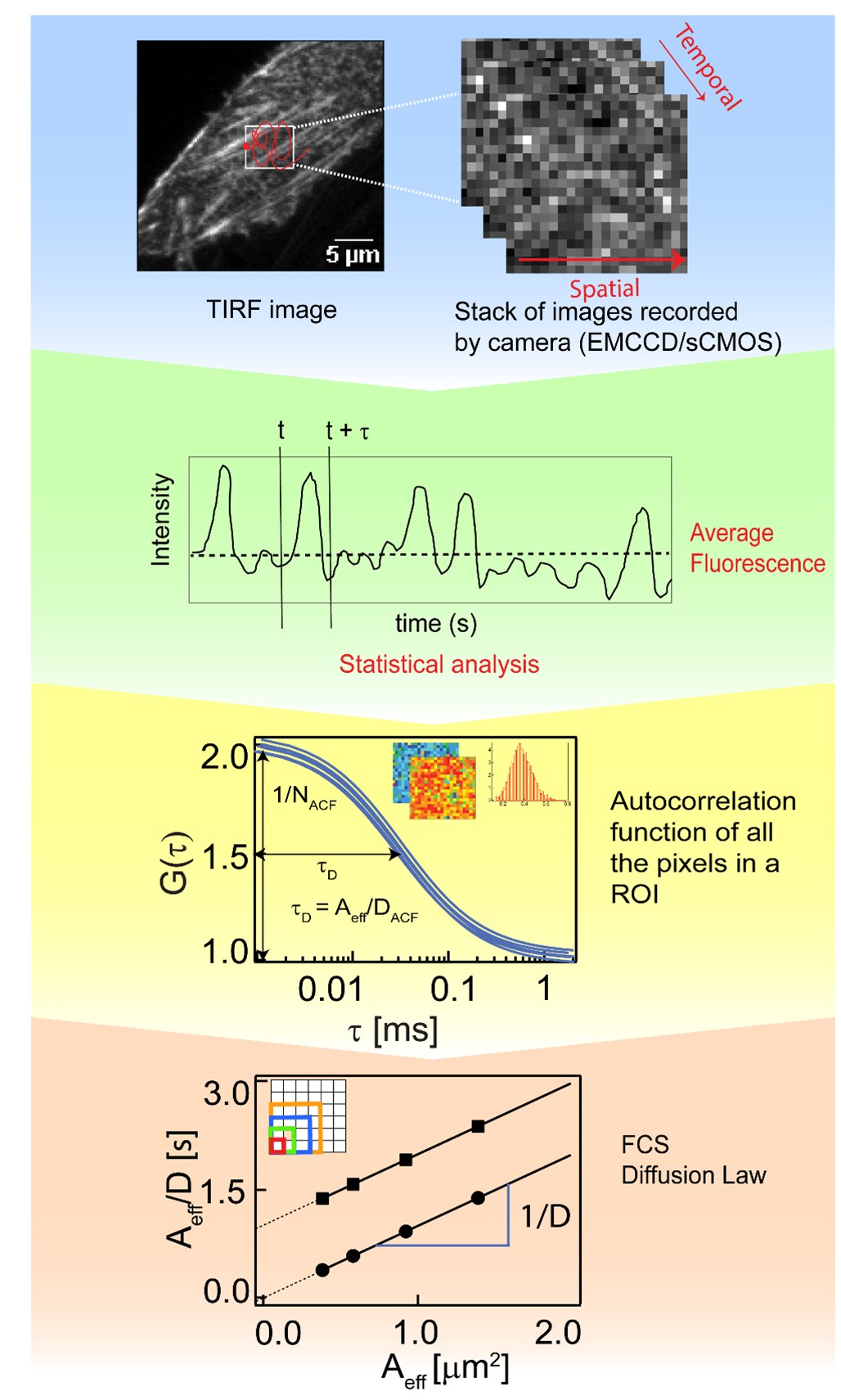
Figure 1 : Workflow of imaging fluorescence correlation spectroscopy. Recording of image stacks with ms time resolution and ~50,000 frames allows the analysis of diffusion dynamics on bilayers by evaluating the correlation of pixel intensities at defined time intervals , the so-called correlation analysis. If, in addition, performed at different length scales, it leads to the FCS diffusion law that will show 0 y-intercepts () for freely diffusing particles, and positive for diffusion with transient domain trapping, providing information on membrane organization. This method enables quantitative analysis of molecular dynamics in solution, 2D cell cultures and living organisms.
ImFCS can be used to determine membrane dynamics and the structural heterogeneities on the plasma membrane (Figure 2). The former has been shown by the measurement of diffusion dynamics of a range of supported lipid bilayers with unique lipid compositions which provided evidence of domain partitioning and hop diffusion in specific lipid bilayers1. The capability of ImFCS to provide information on membrane structure was verified with Atomic force microscopy (AFM) images of bilayers which confirmed ImFCS results2 (Figure 2).
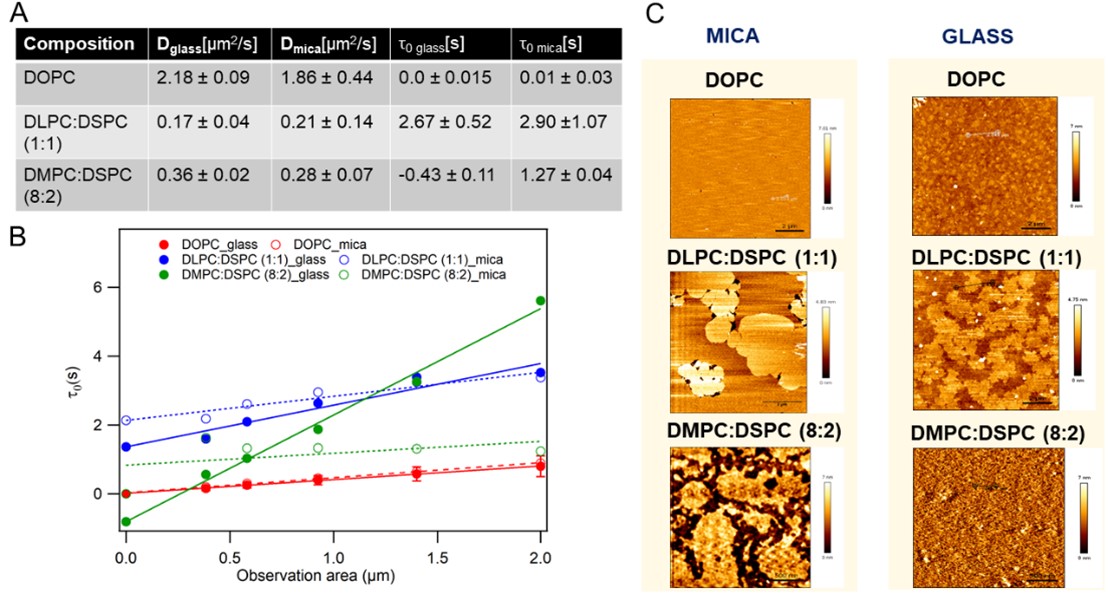
Figure 2 : Direct correlation of the nanoscale organization of supported lipid bilayers (DOPC, DLPC:DSPC (1:1) and DMPC:DSPC (8:2) bilayers) prepared on glass and mica exhibiting diverse domain configurations, as revealed by atomic force microscopy (AFM), with membrane organization as inferred by FCS diffusion law analysis. (A) Summary of diffusion dynamics (diffusion coefficient (D) and FCS diffusion law intercept (t0)) measured on bilayers. (B) Representative FCS diffusion law plots. (C) Representative AFM images of bilayers.
Applications in live cells provided information on the differences in cell membrane structure and lateral dynamics across five mammalian cell types3, showed biophysical differences in the two leaflets of mammalian plasma membranes4, and found correlations between plasma membrane composition and their biophysical properties5.
The workhorse for these measurements was the Andor iXon 860, an EMCCD camera that offers high acquisition rates (up to 3000 frames per second (fps) on small ROIs or 500 fps on the full chip) and an EM gain of up to 300, which helps achieve a sufficient signal-to-noise ratio for FCS measurements. ImFCS has also been performed using the Andor Sona 4.2B-11 sCMOS camera with similar results (Figure 3).
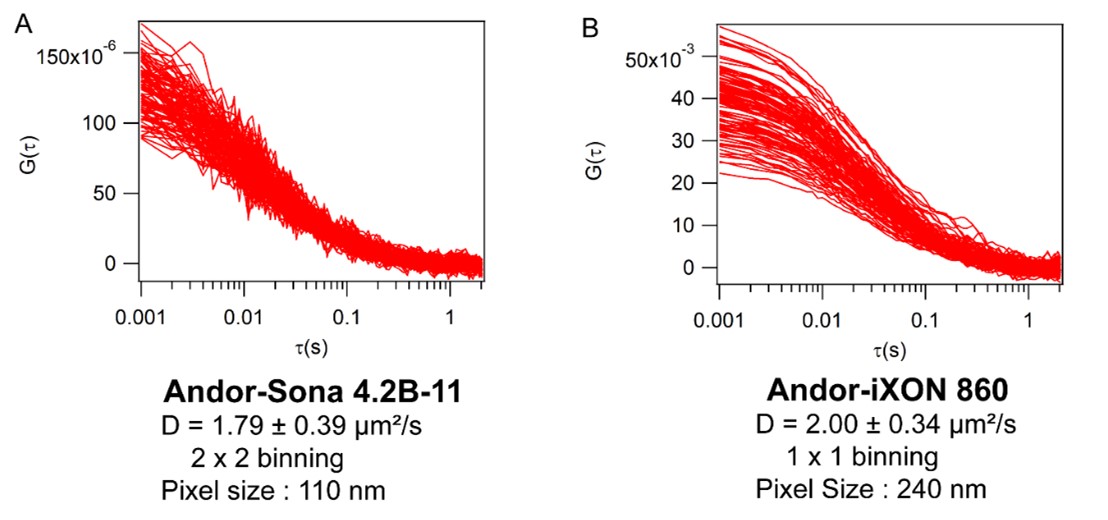
Figure 3: Comparison of Imaging Fluorescence Correlation Spectroscopy data obtained by measuring rhodamine-PE labelled DOPC bilayer. (A) Data obtained using sCMOS Andor-Sona 4.2 B-11 camera; (B) Data obtained using EMCCD Andor-iXon 860 camera.
The imaging fluorescence correlation spectroscopy (ImFCS) open-source software for data acquisition and evaluation is available from the Biophysical Fluorescence Lab webpage along with tutorials on FCS and how it may be applied to study of biological systems.
Date: August 2021
Author: Professor Thorsten Wohland, Department of Biological Sciences, National University of Singapore
Category: Case Study
