Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
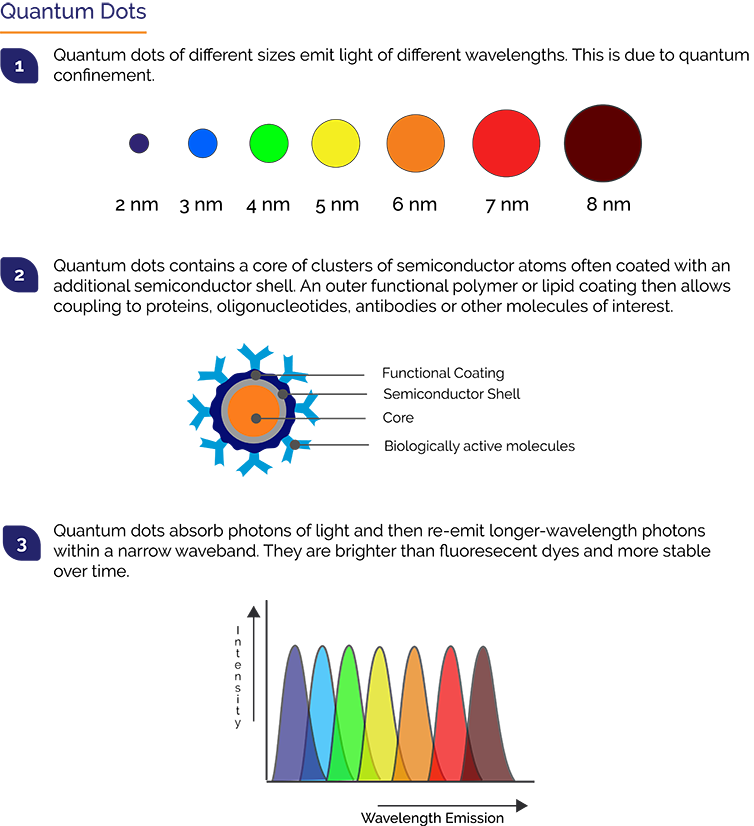
Quantum Dots, also called Qdots or QDs, are semiconductor crystals of between 2 and 10 nm in size. They can emit much brighter fluorescence than conventional fluorescent proteins like GFP, are stable and have a discrete emission spectrum. These optical qualities have made Qdots very useful for single molecule tracking studies in which the typical photon levels are very low. A similar light regime is also present in many virology studies, notably so for live-cell tracking. For live cell tracking the signals from labelled viral proteins are very low making it especially challenging. Techniques for efficient labelling of viruses with Qdots have now been established.
Reoviruses are a group of non-enveloped double stranded RNA viruses that are ubiquitous in the environment. They have a broad host range that covers animals, plants and microorganisms. In humans, reoviruses normally are associated with minor illnesses affecting the gastrointestinal system. Some of this group such as aquareoviruses, are pathogens of commercially important fish stock such as Salmon.
Virus entry into host cells is the first step of any successful infection and therefore a prime target for developing antiviral drug and therapeutic treatments. Viruses use a number of strategies to overcome the membrane barriers such as membrane fusion or endocytosis. Non-enveloped viruses like reoviruses cannot use membrane fusion, and thus generally use endocytosis. Studying viral entry is technically challenging and this process was unknown for this reovirus.
In a recent publication in Frontiers in Microbiology “Real-Time Dissecting the Entry and Intracellular Dynamics of Single Reovirus Particle” Dr Jai Liu, Professor Qi-ya Zhang and colleagues showed that they could apply Qdot-based single particle tracking based microscopy to reveal the infection process of a reovirus in real time. The virus they studied was an aquareovirus: Scophthalmus maximus reovirus (SMReV). Qdot imaging was supported and validated by Biochemical assays and Electron Microscopy.
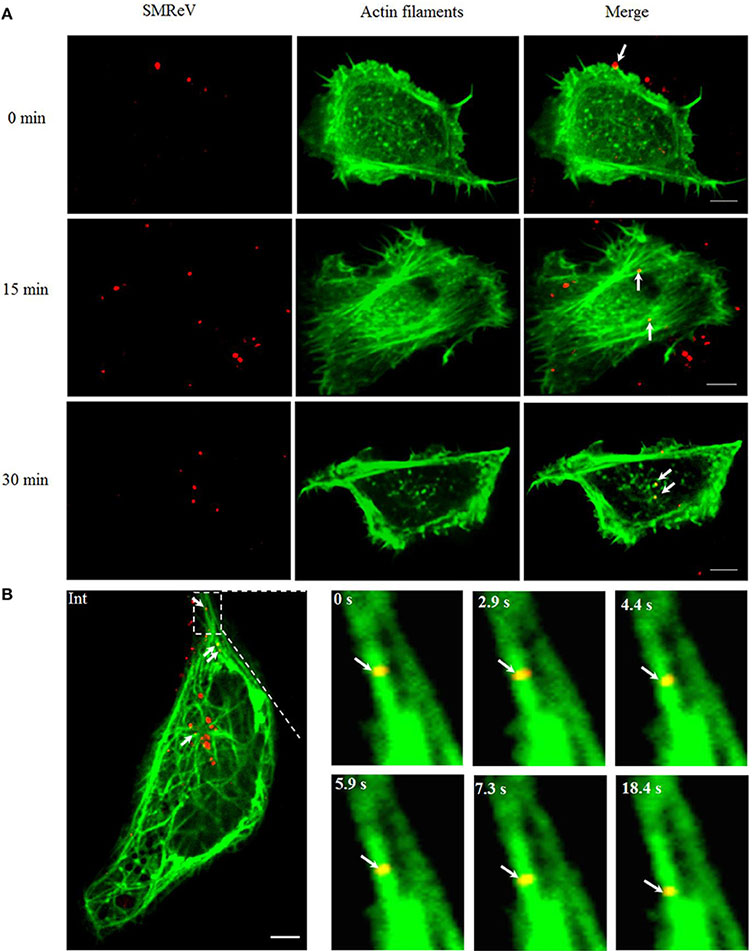
Figure reproduced from Liu et al. Visualization and tracking of virus and cytoskeleton components. Cytoskeleton-dependent movement of SMReV particles in living GCF cells. (A) Time course of SMReV colocalization with actin filaments. QDs-SMReV particles (red) colocalized (yellow) with pRFP-LifeAct (green) labeling actin-rich protrusion at 0 min p.i. (0 min), with long actin filament near the cell periphery region at 15 min p.i. (15 min), and with dot-like actin at 30 min p.i. (30 min), respectively. (B) Snapshots of SMReV particles and microtubule. QDs-SMReV (Int, red) colocalized with pEGFP-MAP4 labeling microtubule (green) showing yellow signals (arrows mark). The enlargement images at different time (0–18.4 s) showed SMReV moved along the microtubule toward the cell interior. Bar: 5 μm.
The results showed that the single membrane-bound reovirus particle can enter the cell within several seconds via nascent clathrin-coated pits. Most of the virions were found to be internalized within the cytoplasm within 30 min of infection. Using specific inhibitors, Liu et al. Then showed that entry of SMReV was dependent on clathrin-mediated endocytosis, and not cavolin-mediated endocytosis. It was possible to continue to track the motion of the internalized single virions labelled with Qdots after entry into the cell. Analysis showed an initial slow and directed motion within the actin-dense areas adjacent to the inner-membrane, and faster motion towards the cell interior. This indicated that SMReV transport was mediated via the cytoskeleton. Further, colocalization using dual-labeling of the virus and cytoskeleton and inhibitor analysis both demonstrated that transport of SMReV was dependent on actin filaments at the cell periphery, and then on the microtubules towards the cell interior. Interestingly, virus particles were also being trafficked and sorted ana packaged via the endosome-lysozome system, through early endosomes to late endosomes, then delivered to lysosomes.
Fluorescent images of the Qdot labelled virions were obtained through an Andor spinning-disk based confocal microscope using 100x objective and an on-line culture system (INUBG2-PI). To detect the resultant signals, an Andor iXon EMCCD camera was used as the imaging camera. This confocal imaging set-up permits the live cell imaging and tracking experiments. The EMCCD camera offers the required sensitivity.
This study has showed that Qdots can effectively be used for tracking live viruses from the initial entry to the host cells through to tracking in association with action and through the microtubules network of the cell.
Research Paper: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02797/full
Date: Aug-20
Author: Dr Alan Mullan
Category: Application Note
