Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Influenza A viruses (IAV) are negative-sense, single-stranded, segmented RNA viruses. IAVs are responsible influenza or “flu” in mammals, including humans as well as birds. Influenza A strains cause seasonal flu epidemics in humans with occasional pandemics causing even higher mortality rates, notably Spanish and Asian Flu pandemics. Influenza viruses also have worldwide economic impacts on livestock for example avian flu (chickens). Influenza A viruses are classified according to the two main envelope antigens hemagglutinin [HA] and neuraminidase [NA] e.g. H1N1.
HA and NA thus have competing activities against sialic acid - being required for binding to the cell and in escape from the cell. A functional balance of these activities is essential for the effective infection of cells - making neuraminidase inhibitors of primary interest as anti-viral treatment. Despite requiring this fine balance in the infection cycle, genetic and morphological variation of viruses play an important aspect of their survival, and therefore their evolution. High mutation rates allow viruses such as IAV evade the immune system and anti-viral drugs. Indeed, the effectiveness of current neuraminidase inhibitors for treatment of IAV in humans is limited.
Electron microscopy and mass spectrometry are useful techniques for investigating the size, structure and composition of viruses but they do not allow for the important variations, and indeed what must be conserved to allow for replicative fitness. Fluorescence microscopy techniques however do allow for studies of single cells over time. This is therefore of interest of virology studies, however it is challenging. Viruses are such as IAV are small in the region of 80-120nm, dwarfed by the host-cells they infect. Labelling components of the virus is therefore difficult without impacting the function of the virus. Additionally, there is only a small amount of label making the signals weak, and below the diffraction limit of light. By using effective labelling strategies and imaging methods such as confocal or TIRF (Total Internal Reflectance Fluorescence), or super-resolution microscopy techniques, aided by high sensitivity EMCCD cameras these challenges can be overcome and are allowing further insights into some of the more complex questions in virology.
Vahey and Fletcher recently reported in Cell, “Low-Fidelity Assembly of Influenza A Virus Promotes Escape from Host Cells” how individual virions could be studied in a population of IAVs, linking their phenotypic variability to their replicative fitness, using confocal and TIRF microscopy.
An important aspect of this study was in the design and use of tags for multiple viral proteins with minimal perturbation of function. Previous studies have used reporter systems with a size of ~25kDa. In this study the tags were small at around 5-10 amino acids in size with small fluorophore making a tag of ~1kD. This is much less likely to affect the function of the virions. Site-specific labelling was used, with a combination of enzymes, probes and FIAsH membrane-permeable dye. Rescued variants had up to 4 tags, making it possible to study populations of IAV virions and how the key envelope antigens affected infection in vivo.
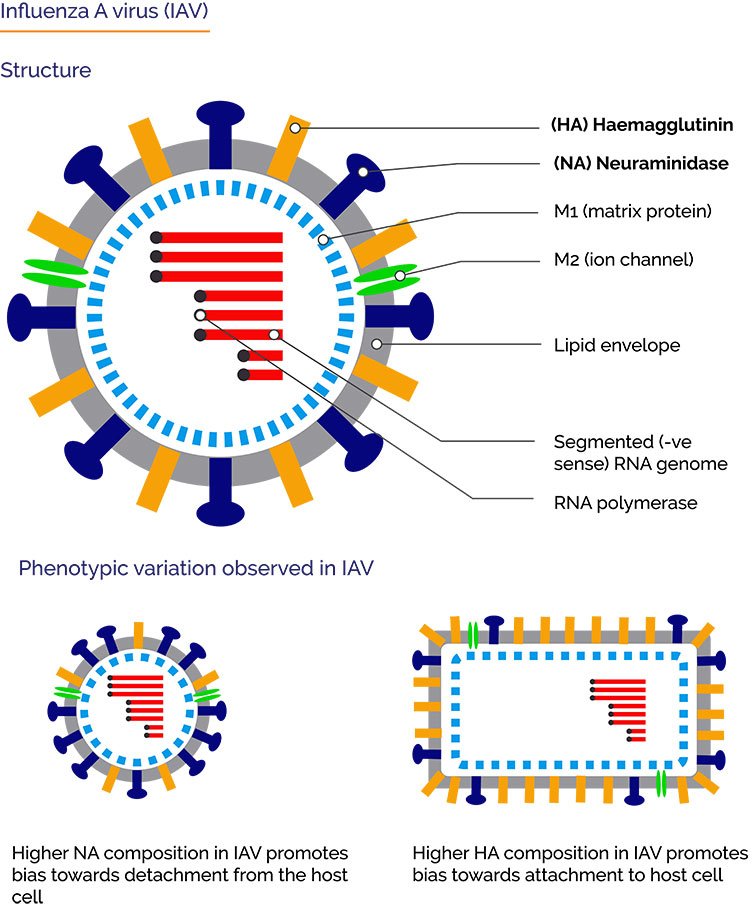 The effect of phenotypic variability on escape of viruses from host cells
The effect of phenotypic variability on escape of viruses from host cellsBy using the labelling technique and the confocal and TIRF imaging techniques described in the study, Vahey and Fletcher were able to make the following observations:
The key finding reported by Vahey and Fletcher was that phenotypic variability in HA/NA composition determines viral release and allows escape from the host cell in response to neuraminidase inhibitors. By adding neuraminidase inhibitor, the virions with a higher composition of NA where able to escape and continue with the next infection cycle. The wide variation in the stoichiometric distributions of HA and NA – at the assembly level, confers IAV an ability to survive in changing environments such as host immune responses, or presence of anti-viral neuraminidase inhibitors. Further work would help further expand our understanding of HA and NA in the infectivity of IAV and potentially lead to novel anti-viral treatments. Beyond this, it would be interesting to see what role phenotypic diversity plays elsewhere for IAV and other viruses.
For confocal imaging, Vahey and Fletcher used a Nikon Eclipse Ti microscope and confocal spinning disk, equipped with an Andor Zyla sCMOS camera, 60X and 100X APO TIRF objectives (1.49 NA), and Andor ILE laser combiner. For TIRF imaging, they exchanged the sCMOS camera for an Andor iXon Ultra EMCCD camera to gain the increased sensitivity necessary for calibrating counts of molecules on the surface of labeled virus. The iXon EMCCD camera provides a higher sensitivity than is possible with sCMOS cameras making it possible to work with small reporter proteins and tags at a single molecule level.
Research Paper: https://www.sciencedirect.com/science/article/pii/S0092867418314557
The Fletcher Lab at Berkeley, has a multi-disciplinary team led by Principle Investigator Daniel Fletcher. The research group has a focus on “visualization and control of biology assembly”. Virus and host cell interactions are therefore just one aspect of a much broader research scope. They develop novel techniques and apply these to in vitro and live cells, to help link biological systems through to processes at the molecular level. Their research covers many of the fundamental aspects of cell biology from biological structures, through to the application of this knowledge, for example potential therapeutic strategies.
Find out more about the fletcher Lab: https://fletchlab.berkeley.edu/
Selected Publications
Date: Aug-20
Author: Dr Alan Mullan
Category: Case Study
