Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Epigenetics is the study of transmissible modifications in gene expression that are not encrypted in the genomic DNA, and for this reason, are named (Epi)genetic - (Above)genetics. Epigenetics is linked with chemical modifications to DNA or chromatin proteins. A common epigenetic modification is DNA methylation. DNA methylation is a repressive epigenetic signature essential for normal development.
The control of transcription is essential for normal cellular differentiation. The Polycomb group (PcG) proteins are required for cellular differentiation during development and act as transcriptional repressors. Interestingly, DNA methylation has an unexpected role in ensuring the correct targeting of Polycomb repressive complexes to the chromatin throughout the genome.
| Glossary | |
| Chromatin | A complex of DNA and protein found in eukaryotic nuclei, which functions to package DNA molecules into more compact, densely packed higher-order structures. |
| DNA methyltransferase | An enzyme that catalyses the transference of a methyl group to the DNA. |
| Epigenetics | Chemical modifications to DNA or chromatin proteins that do not alter the genetic code but can potentially affect gene expression or the coding protein localisation. |
| Heterochromatin | Condensed chromatin which is generally inactive for transcription. |
| mESC | Mouse embryonic stem cells. |
| PCH | Pericentric heterochromatin found at the centromeres of mouse chromosomes. |
| Polycomb | A family of protein complexes that can modify chromatin and promote epigenetic silencing of specific genes. |
|
PRC2 |
One of the two classes of Polycomb-group proteins or (PcG). This complex has histone methyltransferase activity and primarily methylates histone H3 on lysine 27 (H3K27me3) |
The Meehan laboratory (MRC Institute of Genetics & Molecular Medicine, University of Edinburgh) focuses on understanding the epigenetic mechanisms that underpin gene regulation during mouse embryo development. A key research goal is to better understand the relationship between DNA methylation and the Polycomb complex: How do these two systems coordinate to maintain developmental gene silencing?
The Meehan laboratory's previous work revealed that global DNA methylation acts on Polycomb by restricting its deposition to its target sites [1,2]. Further, the group showed that re-programming epigenetic states of mouse embryonic stem cells, leading to a hypomethylated state, induces relocalisation of Polycomb to neighbouring regions. More specifically, the Polycomb components are abnormally deposited at regions of the genome where the pericentromeric heterochromatin (PCH) becomes hypomethylated. The PCH domain contains arrays of major satellite tandem repeats and plays a fundamental role in chromosome dynamics and genome stability, especially during mitosis. The pericentromeric heterochromatin domain is characterised mainly by a repressive signature: the tri-methylation of Lysine 9 on histone H3 (H3K9me3).
The Meehan laboratory's primary goal is to discern if the redistribution of Polycomb components is a passive or active process in hypomethylated mouse embryonic stem cells (mESCs).
Silviya Dimova (PhD graduate student/researcher from the Meehan laboratory) acknowledged that she is fascinated by the consistency by which Polycomb is redirected to PCH in their system. Further, Dimova added, "I am incredibly excited to be able to see the relocalisation in living cells, as so far most of the experiments have been performed in fixed cells."
Dimova used the chromatin reader constructs developed in Tuncay Baubec's laboratory for the live imaging experiments [3]. These constructs were quite helpful since they contain a domain that recognises specific histone modifications. In this case, the epigenetic modification analysed was H3K27me3 (Histone H3, modified on lysine 27 – with three added methyl groups). H3K27me3 is the modification deposited by PRC2 (the Polycomb repressive complex 2).
At the forefront of epigenetics analysis, Dimova and the Meehan group expect to analyse (live) how the Polycomb relocalisation is induced by epigenetic modifications. The plasmid that expresses Cbx7 (a chromatin 'reader' of H3K27me3) fused to GFP and imaging with Dragonfly ultrafast confocal will allow Silviya to be able to follow epigenetic modifications in live cells. (Figure 1).
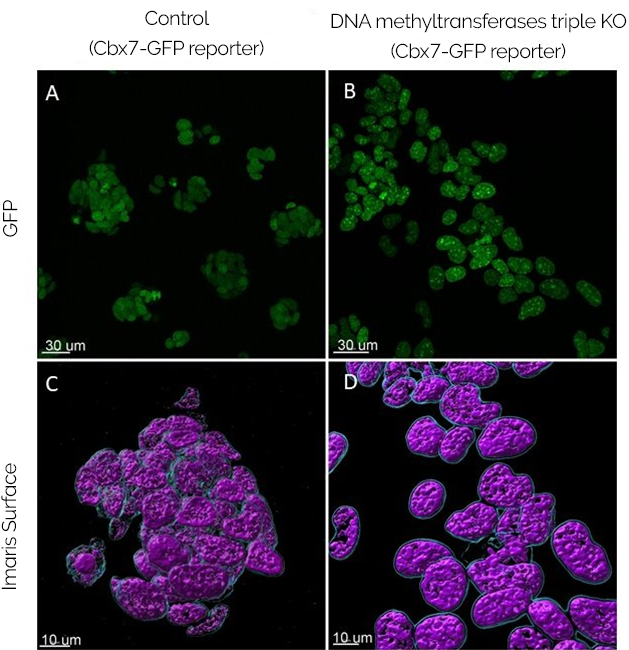
Figure 1 - Initial validation of the results. As can be observed by the picture, the expression of Cbx7-GFP (a chromatin 'reader' of Histone 3 methylation on lysine 27 fused to GFP) is similar between the control and the triple KO cells. Since the expressions are comparable, the initial method is therefore validated. A, B - Live cells were imaged with Andor Dragonfly. C, D – Data analysis was performed by creating surfaces with Imaris 9.6.
The approach taken by the laboratory is to introduce the construct described above in cells that do not have any functional DNA methyltransferases. The plan is to introduce the GFP-Cbx7 construct in DNA methyltransferase triple knockout (TKO) mESCs, cells which have no functional DNA methyltransferases (Dnmts). Hence, the reporter's modification or relocalisation will be caused by the histone modification and not by endogenous methylations. Importantly the relocalisation of the histone modification may also be regulated by DNA methylation.
The initial results are exciting since the GFP-Cbx7 reporter appears as a clustered signal in the DNA methyltransferases KO cells. Therefore, it indicates that there might be a relocalisation of Polycomb away from its regular sites and into these clusters. Being the case, the results will point out to an active relocalisation of Polycomb driven by loss of DNA methylation. This result would mean that some PRC1/2 patterns are dynamically redistributed in mESC in response to altered DNA methylation states, which may also be occurring in early mouse development. Dimova added, "We have yet to confirm whether the clusters formed by the reporter correspond to pericentric heterochromatin."
To analyse the data and perform experiments live, Dimova had to overcome several challenges:
Dimova acknowledged that Andor Dragonfly provided all the essential features that allow live imaging of epigenetic modifications. Dragonfly's combination of speed and sensitivity allows the acquisition of live imaging data in mESC.
"I prefer using the Andor Dragonfly spinning disk confocal microscope to acquire these time-lapse images because it is capable of giving me the resolution I need to observe sub-nuclear events happening in real-time. I am also able to view the progression of my reporter throughout the cell cycle."
Further, Dimova explained that
"Fusion built-in deconvolution is an advantage since it increases the resolution of my images."
As an overall comment on Andor Dragonfly:
"The system is very well optimised and automatic making it extremely easy to acquire large quantities of data suitable for statistical analysis. "
The goal of Dimova is to analyse epigenetic modifications induced in pericentric heterochromatin by Polycomb, and the work is still ongoing. "We found out that a main challenge is the ability to stain the DNA and retain the chromatin – localised signal" (e.g., use a DNA staining agent that would not interfere with the chromatin structure). This result is essential for Dimova's research: to pinpoint the exact localisation of the methylation reporter in relation to the pericentric heterochromatin (PCH).
Pericentric heterochromatin can be observed when staining with either DAPI (or Hoechst). However, because DAPI (and Hoechst) are dyes that intercalate with the DNA, they appear to displace the surrounding chromatin. Using Andor's Dragonfly, Dimova realised that this analysis could only be done by live-cell imaging.
Now, Dimova needs to stain the heterochromatin to analyse the localisation or relocalisation of the reporter. How will the obstacle be overcome? The plan is to use another chromatin – reader (reporter) designed in the Baubec laboratory and stain the pericentric heterochromatin and the reporter GFP-Cbx7 simultaneously. Dimova added that "we are currently in the process of making it compatible with our cells."
As an action movie, we can’t wait to see the next chapters of this story and have molecular knowledge of epigenetic modifications in mESC. Good luck, Silviya and the Meehan laboratory; please keep us posted as the results are collected and analysed!
Date: March 2021
Author: Dr Claudia Florindo, Silviya Dimova, Prof Richard Meehan
Category: Case Study
