Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
The viral capsid provides protection to the viral genome within until it can be delivered into a suitable host cell for replication. Once it has arrived at the correct location within the cell, the capsid structure disintegrates rapidly to release the genetic material in a process known as “uncoating”. In addition to being a protective structure, the viral capsid proteins of HIV have been found to play important roles during infection such as facilitating reverse transcription of the viral RNA and other functions during replication. This means that viral capsid proteins must interact with a range of host-cell proteins and other molecules in different cellular locations and at different times. Despite HIV receiving significant study over many years, this uncoating process and what host co-factors are involved had remained unclear. One of the reasons for this is that the HIV capsids break up on removal from the virus, making it difficult to obtain intact capsids.
The HIV capsid itself is composed of 1000–1500 copies of a single viral protein, CA. HIV capsids are polymorphic with capsids generally conical in shape with a length of ~120 nm. The CA protein subunits may be either hexameric or pentameric and combine to form various heterogenic structures. Therefore many capsids do not form completely closed lattice structures and so are defective. This high degree of polymorphism along with low assembly fidelity are aspects of HIV that makes studying the virus more challenging compared to more regular and uniform icosahedral viruses.
One Research group that has been working to improve our understanding of the uncoating of HIV is the Molecular Machines group of UNSW, Sidney led by Till Böcking. Böcking and colleagues reported a single-molecule analysis based method that allows individual HIV capsids to be visualized – and thus how they disassemble – in real time in their paper, Márquez et al: “Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. In this study, Márquez, Böcking and their colleagues investigated the functionally distinct classes of capsids based on their GFP release kinetics. This permitted the functional heterogeneity for hundreds of capsids within a population to be classified and related to their assembly states. The viral membranes were permeabilized in a controlled manner using a pore-former while the capsid opening traces were monitored. This meant that the unstable assembly states could be observed as they formed, which would otherwise not be possible if isolating capsids in bulk biochemical methods. Viral membrane permeabilization and capsid opening was observed using time-lapse total internal reflection fluorescence (TIRF) microscopy to monitor the release of GFP molecules trapped in these compartments. They also used Cyclophilin A, a host cell protein that binds to the CA, to effectively ‘paint’ the CA lattice and study the lattice as it disassembles.
By using the methods described in the study, it was possible to reveal the capsid uncoating in HIV-1 and interactions with host cell capsid-binding molecules in detail. They showed that the integrity and stability of the HIV-1 capsids varied widely in a population of capsids. Interesting, it was found that the majority of capsids contained defects and/or uncoated immediately after the viral membrane was removed. They went on to define two discrete uncoating events: 1. termed ‘capsid opening’ and another ‘lattice disassembly’ and look at the effect of known cofactors and drugs such as the PF74 (an anti-HIV drug that targets the CA of HIV) on these processes. These studies have helped to resolve conflicting data reported in previous studies for the effect of PF74 on the lattice stability of the capsid.
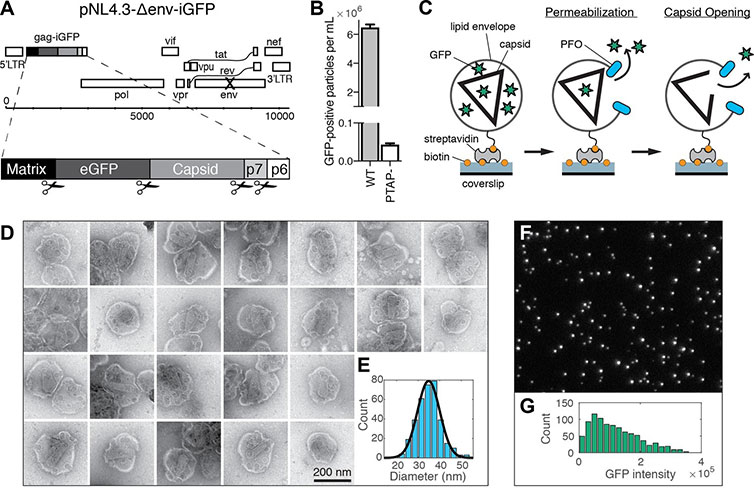 Reproduced from Marquez et al. (A) Map of the proviral DNA contained in the vector pNL4.3-iGFP-ΔEnv. Viral particles with Gag-internal GFP (released as a solution phase marker by proteolysis during maturation) were generated by transfection of HEK293T cells with pNL4.3-iGFP-ΔEnv and psPAX2 in a molar ratio of 1.4:1. (B) GFP-positive particles released from HEK293T cells transfected with either wild type or PTAP motif mutants of pNL4.3-iGFP-ΔEnv and psPAX2. (C) Schematic diagram of the TIRF assay to measure capsid opening. (D) Gallery of negative staining TEM images of viral particles incubated with the pore-forming protein PFO. (E) Distribution of PFO pore diameters (34.8 ± 5.2 nm, mean ±standard deviation, N = 347). (F) TIRF image of immobilized viral particles in a 450 × 370 pixel region of the field of view. (G) Distribution of GFP intensity for each particle in the field of view.
Reproduced from Marquez et al. (A) Map of the proviral DNA contained in the vector pNL4.3-iGFP-ΔEnv. Viral particles with Gag-internal GFP (released as a solution phase marker by proteolysis during maturation) were generated by transfection of HEK293T cells with pNL4.3-iGFP-ΔEnv and psPAX2 in a molar ratio of 1.4:1. (B) GFP-positive particles released from HEK293T cells transfected with either wild type or PTAP motif mutants of pNL4.3-iGFP-ΔEnv and psPAX2. (C) Schematic diagram of the TIRF assay to measure capsid opening. (D) Gallery of negative staining TEM images of viral particles incubated with the pore-forming protein PFO. (E) Distribution of PFO pore diameters (34.8 ± 5.2 nm, mean ±standard deviation, N = 347). (F) TIRF image of immobilized viral particles in a 450 × 370 pixel region of the field of view. (G) Distribution of GFP intensity for each particle in the field of view.
The method reported by Márquez et al. provides an effective means to investigate how the viral capsids interact with the host cell. It could also be possible to apply this more generally to assess the action and effectiveness of potential antiviral drugs that would target the capsid of HIV and other viral pathogens. Since publication, this technique has been applied to a number of other studies.
For these studies a custom built TIRF microscope based around an ASI-RAMM frame (Applied Scientific Instrumentation) with a Nikon 100 × CFI Apochromat TIRF (1.49 NA) oil immersion objective was used. Lasers were incorporated using the NicoLase system (Nicovich et al., 2017). Images were captured on two Andor iXon 888 EMCCD cameras (Andor Technology Ltd). 300 mm tube lenses were used to give a field of view of 88.68 μm × 88.68 μm. A second TILL Photonics TIRF microscope was also used with a Zeiss 100 × Plan Apochromat (1.46 NA) oil immersion objective and solid-state lasers for excitation. A beam splitter allowed for simultaneous dual channel acquisition of fluorescence emission using two Andor iXon 897 Ultra EMCCD cameras for detection with a field of view of 46 μm × 46 μm.
Research paper: https://elifesciences.org/articles/34772
The Molecular Machines research group led by Till Böcking focuses on elucidating the mechanisms of molecular machines in cellular assembly and disassembly processes using a combination of biochemical and biophysical approaches. In particular, they develop fluorescence imaging approaches to visualize the dynamics of these processes at the single-molecule level. Areas of research include: viral uncoating, the assembly pathways of the actin cytoskeleton, visualization of chaperone-mediated processes and understanding the properties of surfaces and materials at the molecular level.Their work draws on approaches from the physical sciences with development of microfluidic imaging devices, surface chemistry approaches and development of automated image analysis software.
Find out more: https://sms.unsw.edu.au/group/molecular-machines
Date: Aug-20
Author: Dr Alan Mullan
Category: Case Study
