Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Traditional light microscopy is limited by the diffraction of light. Consequently, features less than 200 nm apart cannot be resolved. For a significant time microscopy technique development was focused towards improving imaging techniques to allow individual molecules to be resolved. Super-resolution microscopy was developed at the beginning of the 21st century to allow imaging at a resolution beyond the 200 nm barrier (Schermelleh, Heintzmann et al. 2010, Schermelleh, Ferrand et al. 2019). However, super-resolution techniques have limitations, such as limited field of view, limited imaging depth, incompatibility with multicolour experiments, slow acquisition time and high energy illumination intensity requirements. To overcome these hurdles, a few creative research groups went in a different direction and explored what could be done to allow standardised and straightforward nanoscale imaging. Expansion Microscopy (ExM) is an imaging protocol which allows conventional light microscopes to see sub-diffraction limited (<200 nm) or densely packed details which previously could not be distinguished. The development of this new modality of magnification was reported in 2015 by Edward Boyden's team at MIT. While attempting to overcome the difficulties in mapping molecules across the large scales of neural circuits in the brain, Boyden's research group developed a way to magnify the specimen itself, instead of magnifying the emitted signal from the specimen. (Chen, Tillberg et al. 2015).
Table 1- Glossary.
| Glossary: | |
| AcX | acryoyl-X, SE, 6 - (acryoyl amino hexanoïc) ester succinimidylique (crosslinking agent that reacts with amines of proteins and be copolymerized in polyacrylamide matrices) |
| DiExM | differential expansion microscopy |
| DMAA | N,N dimethylacrilamide acid - (nonionic acrylic monomer that can make swellable polymeric particles) |
| ExFish | expansion microscopy fluorescence in situ hybridization |
| ExM | expansion microscopy |
| FP | fluorescent protein |
| GA | glutharaldehyde (crosslinking fixative, penetrate membran more slowly than PFA) |
| HCR | hybridization chain reaction |
| iExM | iterative expansion microscopy |
| IF | immunofluorophore |
| MA NHS | methacrylic acid N-hydroxysuccinimide ester (crosslinking agent that reacts with amines of proteins and can be copolymerized in polyacrylamide matrices) |
| MAP | maximum analysis of the proteome |
| MBAA or BA | N’N Methylenebis (acrylamide) /(bisacrylamide) crosslinking agent that polymerize with acrylamide and creates crosslinks within the polyacrylamide gel) |
| PFA | paraformaldehyde (crosslinking fixative, preserves the secondary/tertiary structures of proteins) |
| ProExM | protein-retention expansion microscopy |
| SDS | sodium dodecylsulfate (anionic surfactant useful to denature and dissociate proteins) |
| SRRF | super resolution radial fluctuations |
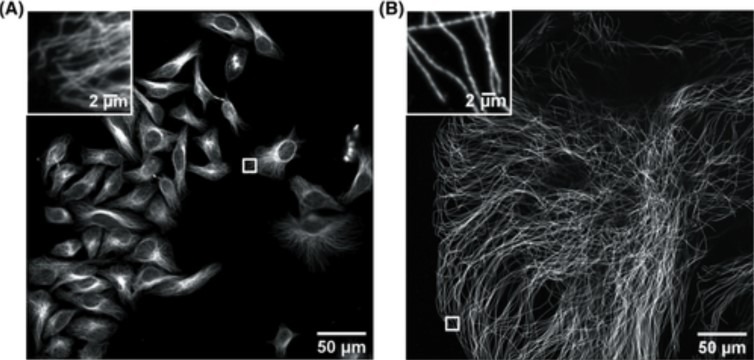
Figure 1. Confocal image of HeLa cells non expanded microtubules (right) and 4.5x linearly expanded microtubules (left). Imaging was performed on an Andor spinning-disk confocal microscope (Dragonfly) with a 40×, numerical aperture (NA) 1.15 water- immersion objective. (A) Confocal image of HeLa cells with immunostained microtubules, imaged at a single xy plane at the bottom of the cells. The inset in the upper left zooms in on the small box at the middle right. (B) Confocal image of a ∼4.5× linearly expanded HeLa cell with immunostained microtubules, imaged at a single xy plane at the bottom of the cell. The inset in the upper left zooms in on the small box at the bottom left. Scale bars in (B) indicate post-expansion scales. Only a fraction of an expanded cell fills the entire field of view. The respective insets display a zoom of the respective small boxes of the full field of view. (Zhang, C., et al, Current Protocols in Neuroscience ,2020).
ExM is a cost-effective sample preparation method which consists of synthesising a dense interconnected web of swellable polymer within a biological specimen. The tissue, within the polymer matrix is expanded and labelled. Once immersed in water, the expansion pulls apart the cellular structures isotropically to create large gaps between each biomolecule. The specimen is isotropically magnified, as a result an effective higher resolution is achievable with a standard microscope. A 4x linear expansion is reported in pure water which means a 64x volumetric expansion. Using Andor spinning disk confocal (Dragonfly) on an expended microtubule Hela cells sample shown in Figure 1 reveals how using expansion microscopy protocols, researchers can see previously unseen details in their samples. Nevertheless, care must be taken when preparing expanded samples; successful implementation of the expansion microscopy protocols relies on:
Table 2- Comparison of Expansion microscopy protocols. See the glossary above for the abbreviations
| Protocol name | ExM (Boyden Lab) | ExM (Vaughan Lab) | ProExM (Boyden Lab) | MAP (Chung Lab) | ExFiSH (Boyden Lab) |
| Hydrogel | acrylamide + sodium acrylate +++ MBAA | acrylamide + sodium acrylate +++ | acrylamide + sodium acrylate +++ | acrylamide +++ sodium acrylate + BA | acrylamide sodium acrylate MBAA |
| Linking agent | acrydite | MA-NHS GA | acryloyle-X (AcX) | paraformaldehyde acrylamide | LabelX (AcX + Label-IT amine) |
| Disruption agent | proteinase K | proteinase K | proteinase K | SDS | proteinase K |
| Disruption type | digestion | digestion | digestion or gentle disruption | denaturation dissociation | digestion |
| Expansion factor (linear) | 4.5 | 4.0-4.2 | 4 | 4 | 3.3 |
| Resolution | 70 nm | 65 nm | 70 nm | 60 nm | / |
| FP preservation | no | yes | yes (50% intensity) | no | no |
| IF staining | no | yes | yes | yes | no |
| Sample | cells, brain tissue | cells, brain tissue | cells, brain, pancreas, lung, spleen tissues | cells, brain, spinal cord, lung, heart, liver, kidney, intestine tissues | cells, brain tissue |
| Target | proteins | proteins & DNA | proteins | proteins & saccharides | RNA & DNA |
| Pros comments | first method reported | conventional fluorophores organelle level | conventional fluorophores post-expansion labelling | post-expansion labelling whole organ level multiplexed staining | 3D imaging post-expansion FISH multiplexing & HCR amplification |
| Cons comments | complex protocol no standard fluorophores | pre-expansion labeling only fluorescence loss | incomplete homogeneization fluorescence loss | incompatibility w/ FPs preparation lost after 3 days | / |
| Reference | Chen et al., 2015 Science | Chozonski et al. 2016 Nature Methods | Tillberg et al., 2016 Nature Biotech | Ku et al., 2016 Nature Biotch | Chen et al., 2016 Nature Methods |
| Variant | iExM | x10 | x10; DiExM | uExM |
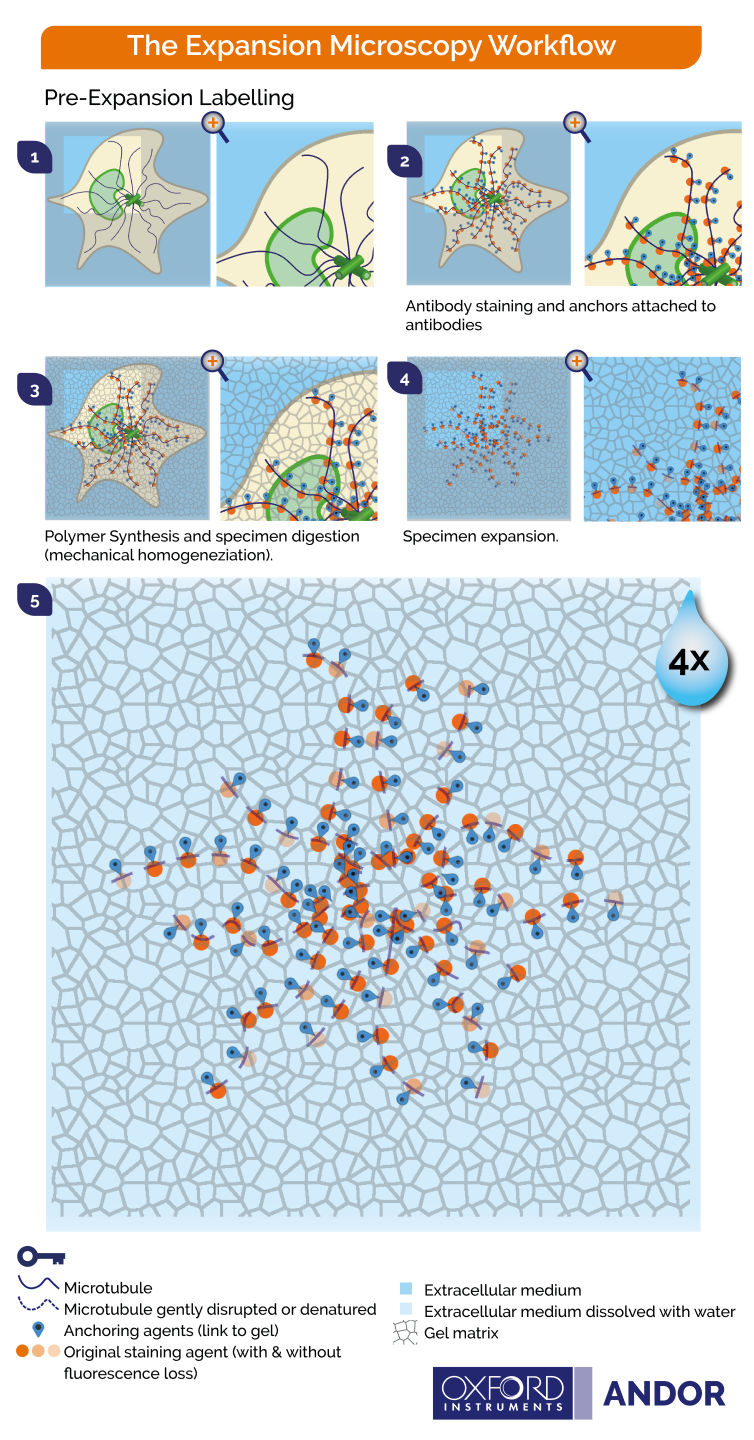
Figure 2- Principles of pre-expansion microscopy. 1) Cells are fixed 2) Labelling by immunostaining is performed on the samples and biomolecules are covalently anchored to the gel 3) During gelation the specimen is immersed in a monomer solution and the chemical network is formed. 4) During homogenization, the specimen structures are chopped by enzymatic digestion to ensure that the organization is kept intact. 5) Upon water immersion, spontaneous expansion occurs. A 4-fold linear expansion is reported in pure water (64 volumetric expansion).
There are 2 main approaches for expansion microscopy:
The graphics shown in Figure 2 and Figure 3 represent the protocols in which cells and tissues are expanded up to 4x. The 200 nm diffraction limit of light sets a boundary on what can be distinguished as separated by light microscopes. Since the linear expansion achieved is a factor of 4, using these protocols, biomolecules that are separated up to 50 nm will be visible under the light microscope.
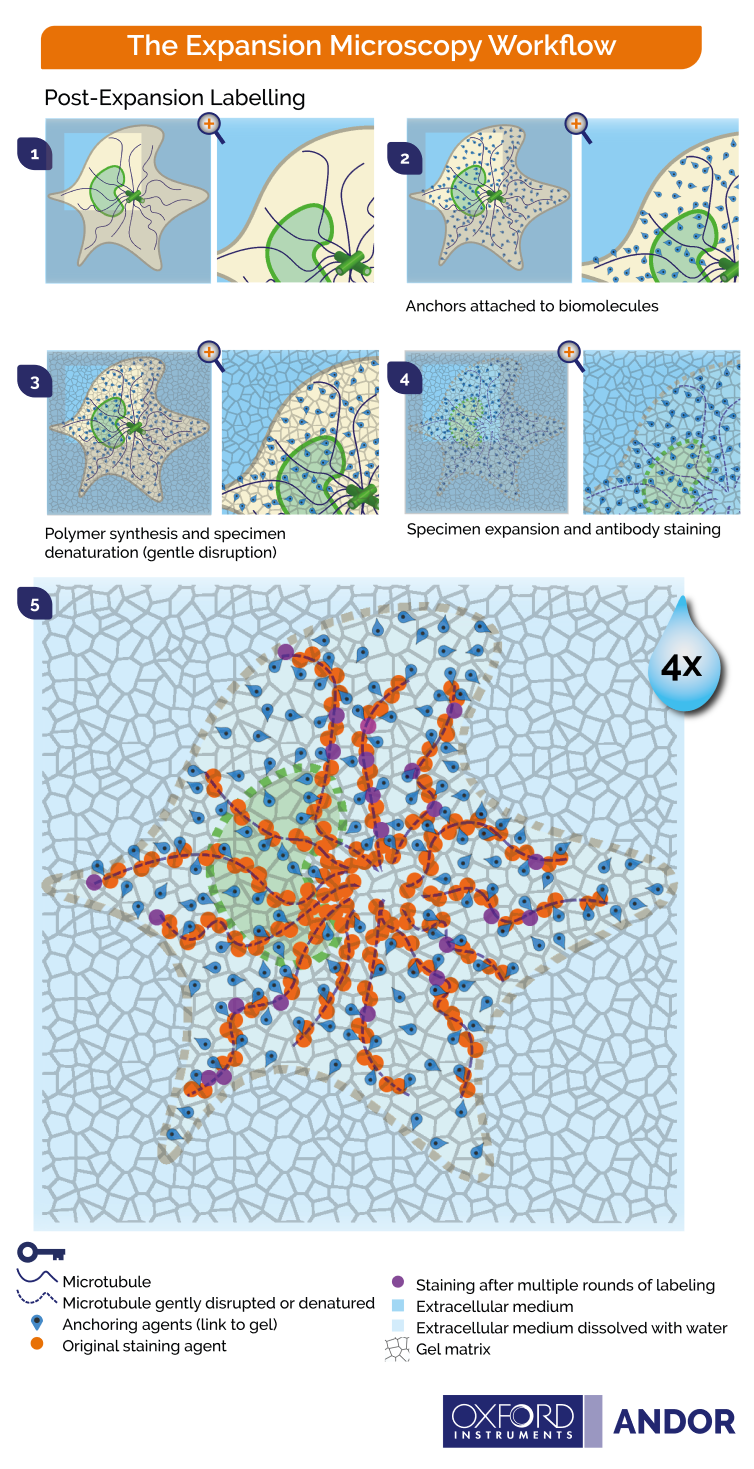
Figure 3- Principles of post-expansion microscopy 1) Fixation of cells 2) Covalent anchoring of endogenous proteins to the gel 3) Hydrogel embedding with high polyacrylamide concentration 4) Denaturation and dissociation of non-crosslinked proteins 5) Expansion upon water immersion 6) Post-expansion immunostaining possible in multiple rounds.
Through expansion, the fluorescent signal is also isotropically expanded. As a result, an effective higher resolution is achieved. The key steps for expansion microscopy protocols are:
(see also Figure 2 and Figure 3):
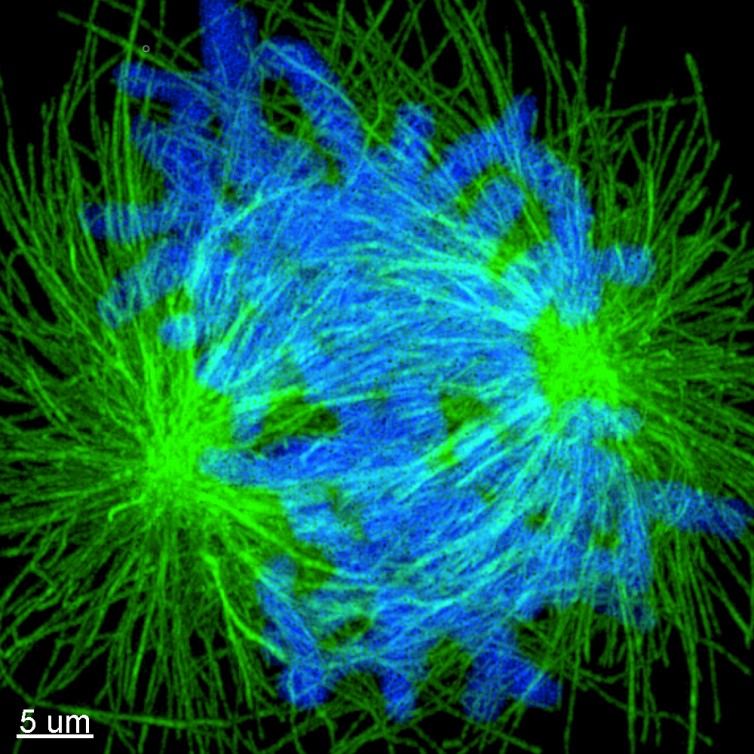
Figure 4. Image of expanded mitotic cell. Maximum projection images of dividing cells stained for tubulin (green) and DNA (Blue). Cells where labelled using the post-expansion protocol, and image with Andor Dragonfly. Sample courtesy of Joshua C Vaughan (University of Washington).
Since its development, several different Expansion Microscopy protocols have been published. Currently, there are variations and improvements which cover a wide range of applications. Below we present a table summarizing the main protocols for expansion microscopy.
In summary, two main strategies of expansion microscopy co-exist: the pre and the post-expansion labelling strategies (figures 2 and 3), and the different protocols are adapted to specific cellular structures. When starting on expansion microscopy imaging, care must be taken to choose the correct protocol to visualize the desired subcellular structure. It might be necessary to optimize a protocol to better suit the experimental conditions; researchers should not forget to design the proper experimental controls to demonstrate the isotropic expansion of the structure to be analysed.
Learn more about Andor’s product solutions for Expansion Microscopy in our solution note - Dragonfly the ideal multimodal imaging platform for Expansion Microscopy.
Bibliography:
Date: Sep-20
Author: Dr Sébastien Bellow & Dr Claudia Florindo
Category: Technical Article
