Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
The first step in the infection cycle of a virus is the attachment to complementary cell receptors on the surface of a suitable host-cell. For the human immunodeficiency viruses HIV-1 and HIV-2 the primary cell receptor is CD4 found notably in T-helper cells. T-helper cells perform a range of roles as part of the immune system and the depletion of these cells due to HIV infection allows other opportunistic pathogens to exploit the suppressed immune system. The main steps during attachment of an HIV virion with a CD4+ cell start with the Envelope spike glycoproteins gp120 of the HIV virus docking and binding to a CD4 receptor of the host cell. Following this, additional co-receptors CCR5 and/or CXCR4 are also involved in binding. Eventually, the viral envelope fuses with that of the cell membrane in a process mediated by the virus gp41 transmembrane protein, and the HIV capsid is released into the host-cell. Reverse transcription then converts the viral RNA into viral DNA. The viral DNA integrates into the host cell DNA and replication of the virus then proceeds.
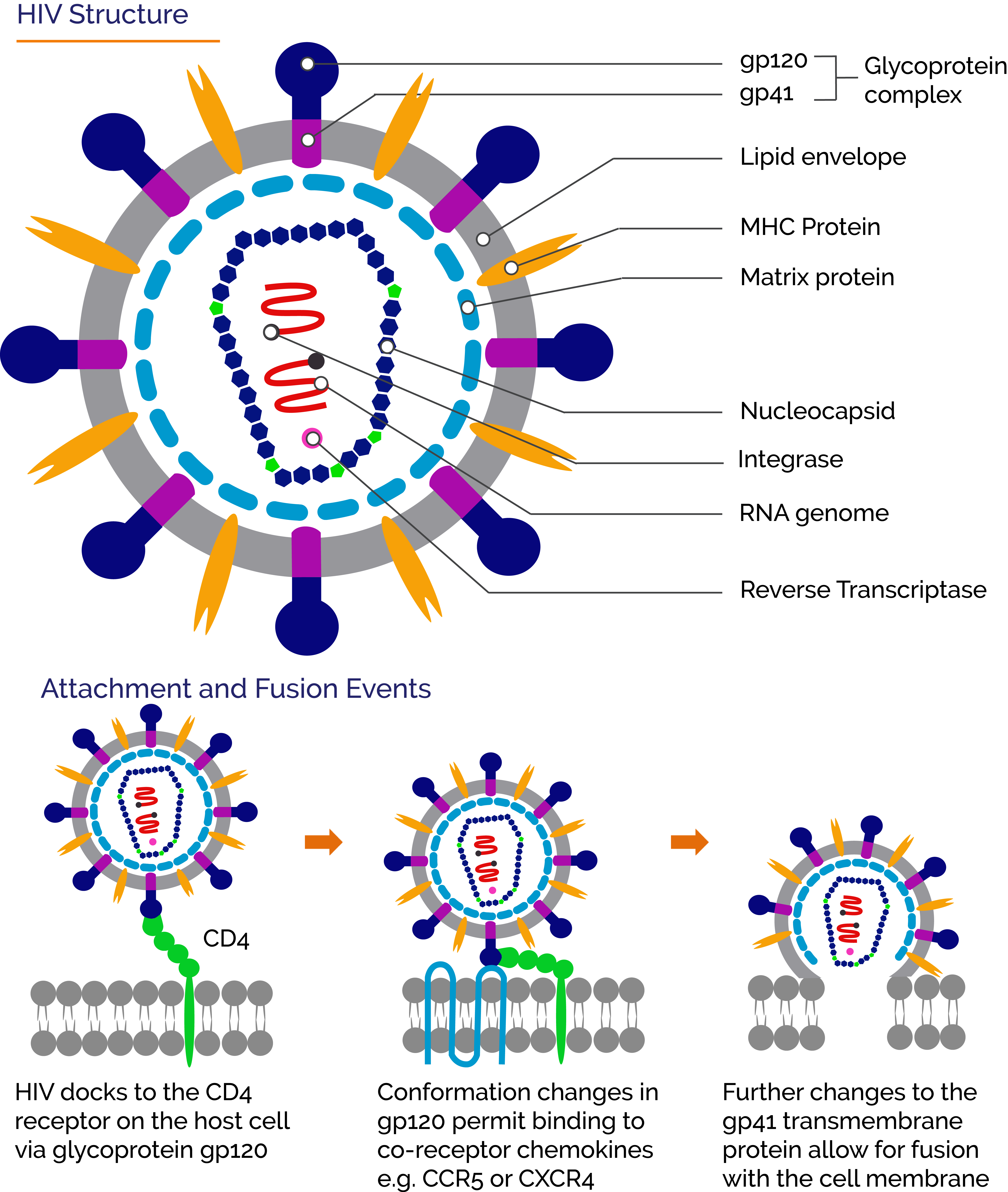
The study of attachment and fusion events of the virus cycle are of interest because they are potential targets for anti-viral treatments. Although anti-viral drugs targeting attachment and fusion events of HIV are available, it is still important to study these processes in more detail. HIV has a high degree of heterogenicity and variation within a single patient. Additionally by integrating into the host cell DNA it establishes latent infections. These factors make it hard to tackle from a clinical perspective. Current treatments utilize a number of drugs that target different points within the virus cycle, greatly reducing the viral load and allowing the population of CD4+ cells such as the T-helper cells to rebuild. Studying the dynamic steps from attachment through to fusion with the host-cell membrane in live cells is very difficult. Techniques such as Electron Microscopy and crystallography provide a wealth of structural information and a highly resolved snapshot of events. However, they do not provide any temporal information for live cells that allows for a greater insight into the processes involved over time.
Recently developed labelling and fluorescence microscopy techniques do provide ways to study viruses and their interactions with live cells over time. Components of the virus and the cell can be fluorescently labelled and interactions can be observed using microscopy techniques such as confocal, FRET, TIRF (Total internal reflection microscopy) or super-resolution. In a recent paper “A dynamic three step mechanism drives the HIV-1 prefusion reaction”, the group of Maro Iliopoulou, Sergi Padilla-Parra and co-workers used fluorescence microscopy spectroscopy and imaging techniques to study the attachment and binding events leading to membrane fusion in real time of laboratory and primary strains of HIV-1. In this study they combined real-time single virus tracking (SVT) with Number and Brightness (N&B) analysis. To visualize these processes and support the quantitative experiments they used TIRF, super-resolution localization microscopy (dSTORM), and also Multicolour TIRF-dSTORM imaging. High sensitivity Andor iXon Ultra 897 EMCCD detectors were used to capture the weak signals involved.
By using this combination of techniques they were able to observe the steps of attachment through to fusion effectively and provide insight of the intermolecular dynamics of these important steps, comparing both laboratory strain: X4 tropic HIV-1 (HXB2) and primary isolate R5 tropic HIV-1 (JR-FL). As part of this Iliopoulou et al proposed a three-step models for the prefusion process based on the Fluorescence Fluctuation Spectroscopy measurements. For HIVHXB2 this included a sequence as follows: 1. Envelope to CD4 binding creating an asymmetric pre-hairpin intermediate, 2. Formation of a CD4 hexamer and co-receptor CXCR4 dimerization and 3. The disassembly of the CD4 hexamer into a CD4 tetramer. For HIV-1JR-FL however a different stoichiometry was involved after envelope to CD4 binding with recruitment of other CD4 receptors and co-receptors before fusion. The reported findings for these different processes indicate potential routes to investigate anti-viral drugs that target formation of the required receptor and co-receptor oligomerization complexes to allow fusion to proceed.
Multicolor TIRF-dSTORM microscopy was used to support the data allowing multiple components to be visualized with high resolution during these steps. By incorporating FLIM-FRET experiments it is also possible to quantify differences in fluorescence intensity of labelled components and study co-localization as well as the dynamics of binding events.
Application of fluorescence imaging such as single molecule and super resolution microscopy to studies of HIV is relatively new. The study of Iliopoulou et al is an example of how single molecule imaging approaches can be applied in live cells and used alongside quantitative spectroscopic measurements to provide both spatial and temporal analysis of complex processes. Such approaches allow for greater insights into challenging areas of study such as the dynamics of attachment and fusion. Labelling of virus proteins and lipids without perturbation of function is another area were further developments will help to improve our understanding of all aspects of the complex virus-cell interactions during infection.
dSTORM Super Resolution Imaging- Zeiss Elyra TIRF system configured with dual Andor iXon Ultra 897 EMCCD cameras. “Nanoboosters” were used to reduce photobleaching while allowing fluorophore blinking that is required for dSTORM.
Fluorescence Resonance Energy Transfer – Fluorescence Lifetime Imaging microscopy (FRET-FLIM) in live cells- Leica SP8-X-SMD confocal microscope with a 63×/1.40 numerical aperture oil immersion objective. FRET detection was based on time domain FLIM experiments using a Time-Correlated Single Photon Counting (TCSPC) system operated by a PicoHarp 300 module attached to the confocal microscope.
Virus Tracking - Virus tracking was measured and analyses using the tracking module of Imaris. This software provides highly effective tools for tracking fluorescently labelled molecules and viruses over time.
Research Publication: https://www.nature.com/articles/s41594-018-0113-x
Date: Aug-20
Author: Dr Alan Mullan
Category: Case Study
