Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
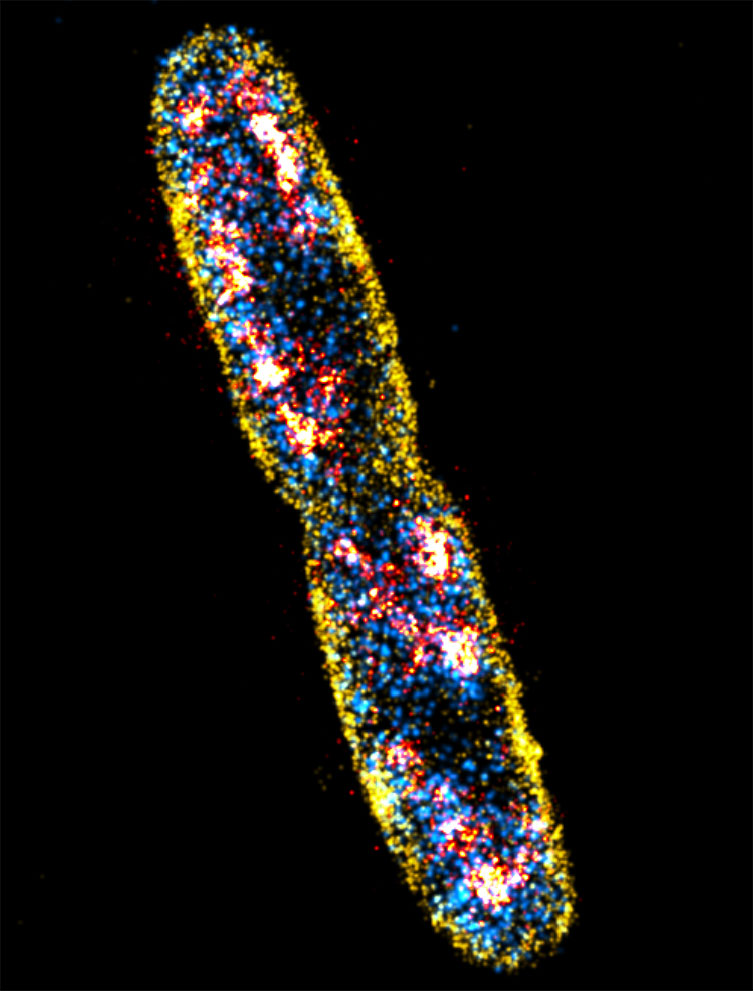
Figure 8: Sequential PALM (RNAp), PAINT (membrane) and dSTORM (DNA) imaging of E. coli cells taken with an Andor iXon EMCCD camera. Scale bar is 1 μm. Courtesy of Christoph Spahn, Ulrike Endesfelder & Mike Heilemann, Institute of Physical and Theoretical Chemistry, Goethe-University Frankfurt
E. coli has been a key model organism from the very earliest work on molecular genetics and continues to play an important role to this day. Much of our understanding of the fundamental concepts of molecular biology such as replication, gene expression and protein synthesis have all been achieved through studies of E. coli.
The E. coli genome is relatively small, 4.5 to 5.5 Mbp [1] and simple when compared to our own (nearly 3 billion bp). Most strains of E. coli are harmless in contrast to the more headline grabbing strains such as 0157 or Verocytotoxin-producing E. coli (VTEC). Strains such as K12 are easily cultured with simple nutritional requirements and have a short generation time as low as ~20 minutes. Mutants are easily obtained using well established methods and screening techniques, which, has enabled many biochemical processes to be linked back to the molecular genetic level. Current research areas of interest for E. coli include acting as a vector, a host for genetic elements and synthesis of proteins of interest.
Recent research in E. coli uses modern techniques such as Super resolution [2] and Andor’s SRRF-Stream, which can provide a boost in resolution to 50-150 nm making it especially useful for the study of E. coli and other bacterial cells.[3]
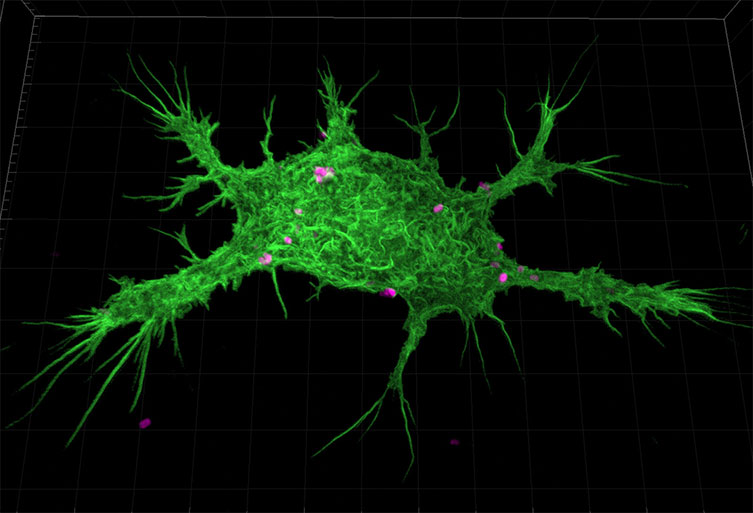
Figure 9: Murine Macrophage stained with AlexaFluor488 Phalloidin phagocytosing E.coli bioparticles labelled Alexa fluor 594. This image was captured with a Zyla 4.2 at 100x magnification with 40 µm pinhole.
References:
Learn More from the Model Organism Series
Date: February 2019
Author: Dr Alan Mullan & Dr Aleksandra Marsh
Category: Application Note
