Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Featured Lab: The Chen Lab, Genome Institute of Singapore
As cells develop and function in tissues, many changes occur not just at the level of RNA transcription, but also at the larger level of spatial organisation of cellular gene expression within the tissue. Studying these changes to RNA profiles at the single-cell level can thus provide a wealth of information about the status of the tissue. Changes to normal RNA profiles also occur in abnormal conditions such as in the development of tumour tissue. Therefore, study and characterisation of this transcriptomic information can be widely applied in areas such as oncology, neuroscience, behavioural studies, drug screening, and disease therapy. One of the research groups that has been advancing research in the area of spatially-resolved transcriptomics is the Chen Lab at the Genome Institute of Singapore.
There are a number of strategies and variants thereof for studying RNA profiles (i.e., the spatial organisation and relative abundance of RNAs) within cells and tissues. These include: multiplexed single molecule fluorescence in situ hybridisation (smFISH), single-cell in situ sequencing, and spatial RNA sequencing. These techniques have different benefits and drawbacks, but they all aim to provide as complete a profile of the cellular transcriptome as possible in intact tissues. In the case of multiplexed FISH, this is achieved by using oligonucleotide probes to detect different RNA species in their original locations. Such techniquesmake it possible to uncover subtle yet important relationships that may not otherwise be apparent. Learn more about using confocal microscopy for Multiplex In Situ Hybridization.
One of the main challenges to applying Multiplexed FISH for RNA profiling of complex tissue samples is that it becomes difficult to detect individual RNA molecules. Complex tissues contribute to the background via autofluorescence while absorption and scattering further attenuate the already low-level signal from the labelled probes. Another problem stems from the non-specific binding of the oligonucleotide probes which not only raises the noise floor but also contribute to false positive signal. Many studies have attempted to amplify the signal against the tissue background. However, the issue that remains is that non-specifically bound probes are also amplified and the false-positives subsequently remain. This is a particular problem for using multiplexed FISH for RNA profiling due to the large number and diversity of probes involved, which make it difficult to optimise for all probe lengths and melting temperatures.
Tissue clearing offers a way to remove cellular proteins and lipids that contribute to non-specific probe binding. One of the limitations of tissue clearing is that it does not address non-specific binding of probes to non-target RNAs. A different approach to tackle the problem is by using “split RNA probes”. A split probe will only generate a fluorescence signal when the two halves of the split probe pair both successfully bind in close proximity. This would significantly reduce the number of false-positives arising from non specifically bound probes and address the challenges to successful application of multiplexed FISH to tissues.
Recently Goh et al., 2020 presented such a technique called “Split-FISH” in their paper “Highly specific multiplexed RNA imaging in tissues with split-FISH”. The team at the Chen Lab reported that Split-FISH allows for RNA profiling in a wide range of tissues.
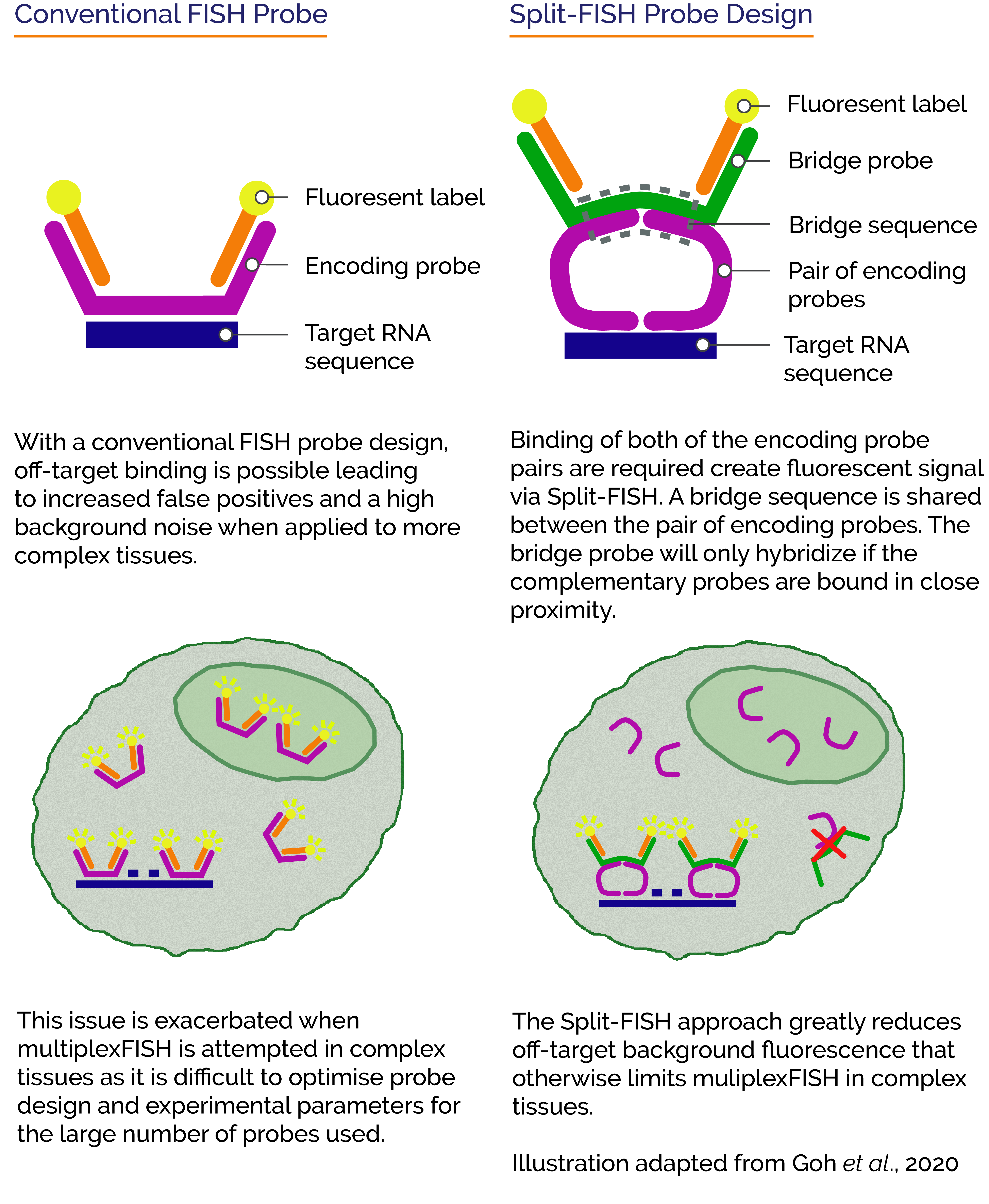
Using the split probe concept, they achieved highly specific labelling of 317 genes, reduced off-target background fluorescence and false-positives within uncleared tissues. They went on to demonstrate the effectiveness of the technique by revealing spatial distributions of the various transcriptomes of individual cells within mouse brain, kidney liver, and ovary tissues.
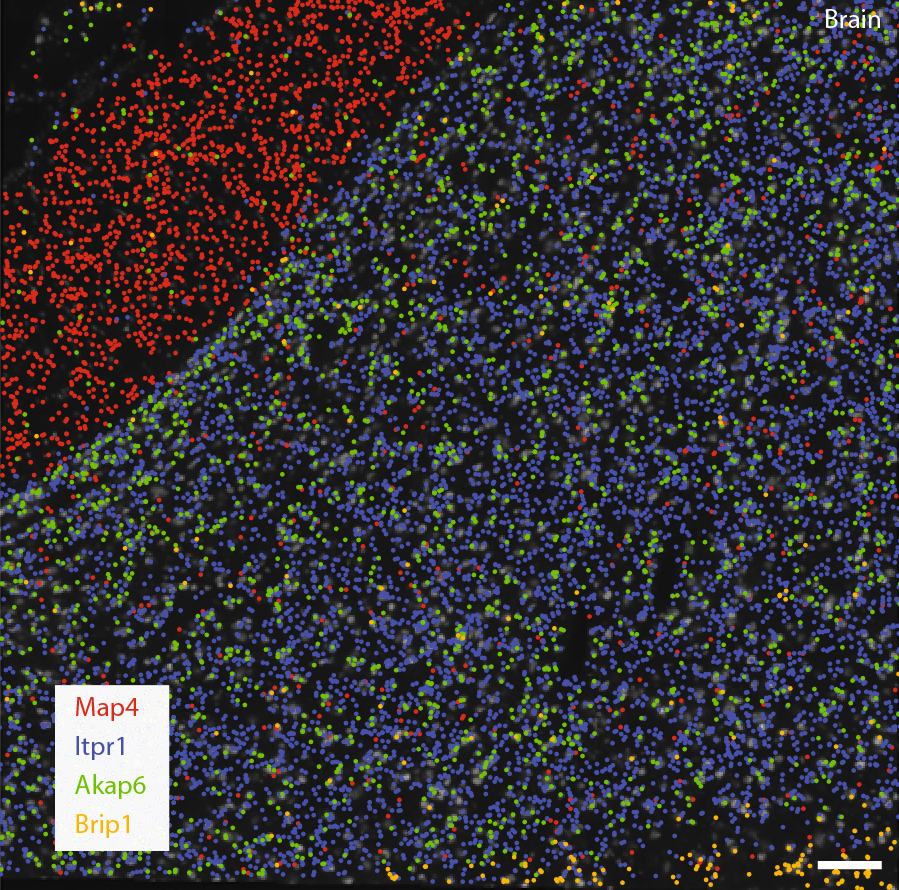 Decoded transcript locations of selected genes are overlaid on stitched images taken with Sona 4.2B-11. Scale bar, 100 μm. In this example, the differential localisation of transcripts in regions with (Itpr1) and without (Map4) cell bodies can be observed in mouse brain tissue. (Goh et al., 2020)
Decoded transcript locations of selected genes are overlaid on stitched images taken with Sona 4.2B-11. Scale bar, 100 μm. In this example, the differential localisation of transcripts in regions with (Itpr1) and without (Map4) cell bodies can be observed in mouse brain tissue. (Goh et al., 2020)
Detectors used for RNA profiling studies should have high sensitivity and low noise to allow detection of the fluorescently labelled RNA probes which may be relatively dim, especially when working with complex tissues. To study larger areas of tissues or fields of cells, a large field of view is also highly beneficial. Since a high throughput for large image datasets is often necessary, it is important to have an imaging camera that runs at high speeds.
For the multiplexed FISH and co-localisation experiments, the Chen Lab used a custom-built microscope based around a Nikon Ti2-E body (Nikon CFI Plan Apo Lambda ×60 1.4 NA oil-immersion objective). A Sona 4.2B-11 back-illuminated sCMOS camera was used as this provided the necessary combination of high sensitivity, speed, and widest available field of view.
For other imaging experiments, a custom-built microscope was based around a Nikon Ti-E body (Nikon CFI Plan Apo Lambda ×100 1.45 NA oil-immersion objective). An iXon Ultra 888 EMCCD camera was used, which provides higher sensitivity, but with slower speeds and a smaller field of views.
Researchers at the Chen Lab are interested in how 'in situ omics' may be applied to answer some of the most complex questions in cell biology and disease processes. 'In situ omics' data can be used to identify and profile molecularly defined cell types, gene networks, and cellular signalling events within their spatial context. This makes it a very effective approach to understand tissue biology and the underlying complex, yet subtle, changes that occur within cells due to disease. To conduct their research, the Chen Lab use some of the latest microscopy tools such as the Dragonfly multimodal confocal system, micro-fluidics systems, and next-generation sequencing systems. Additionally, a key part of their work is in developing computational software tools to help with the analysis of the genomic and transcriptomic information at the single-cell and tissue level.

Find out more about the research of the Chen Lab at: Chen Lab @ GIS (khchenlab.github.io)
