Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Spatial transcriptomics (or Multiplexing) in cell biology is the unveiling of several (Xn) RNAs in its 2D or 3D biological context. Multiplexing has become a hot topic in neurosciences, oncobiology, disease target diagnostics, development, behavioural studies, etc. Several techniques have been developed to allow spatially resolved transcriptomics (also known as spatial genomics or multiplexing); examples of such are FISSEQ, instaSEQ, osmFISH, STARmap, MERFISH and seqFISH.
The advantage of spatial transcriptomics is its ability to deliver the spatial and sequence information of multiple gene products. In fact, up until recently, RNAs could be sequenced in bulk, or at the single-cell level, but the tissue environment information (the biological context) was lost. Nevertheless, understanding where genes are expressed and their surrounding environment, or microenvironment, might reveal the molecular basis of development, brain function, neurodegenerative diseases and cancer.
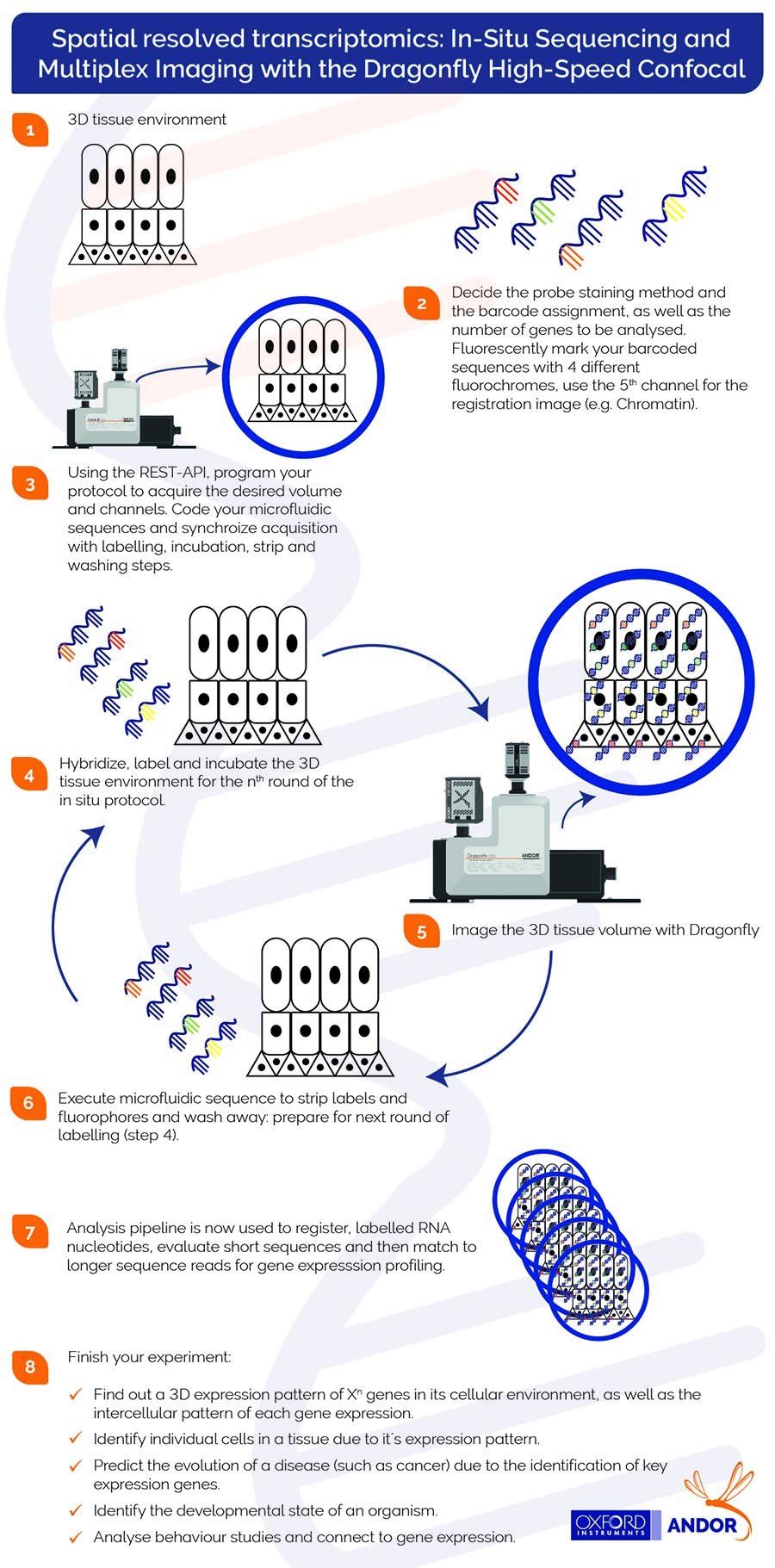
Multiplexing hybridisation experiments proceed with several rounds of hybridisation and detection. These rounds are executed sequentially in the same sample. In order to be able to stain (Xn) molecules, the probes must be removed, and the sample needs to be hybridised again, this can be achieved by attaching a microfluidic system to the Dragonfly Spinning disk confocal system.
Mechanistically, fluorescent probes label the hybridised RNA molecules, the image data is acquired (typically a volumetric montage is scanned), and the probes are washed away. After each image dataset is acquired, a “strip and wash” step is followed by another hybridisation round. This procedure is repeated N times and results in large volumes of encoded image data. In the infographic below, we explain the sequential steps required to acquire multiplex imaging with the Dragonfly confocal.
Date: Oct-20
Author: Dr Claudia Florindo
Category: Solution Note
