Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
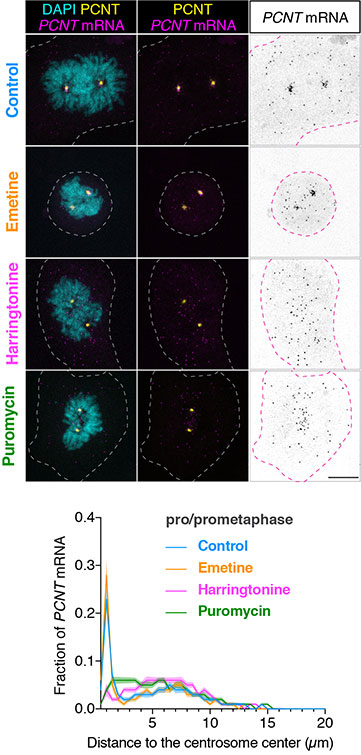
HeLa cells were synchronized by a double thymidine block and treated with DMSO vehicle (Control), emetine, harringtonine, or puromycin before anti-pericentrin (PCNT) immunostaining and PCNT smFISH. Representative confocal images and quantification of the PCNT mRNA distribution are shown for each condition. The researchers quantified the distribution of PCNT mRNA in cells by measuring the distance between 3D rendered PCNT smFISH signals and the center of the nearest centrosome. The fractions of mRNA as a function of distance to the nearest centrosome were then plotted as mean (solid lines) ±95% CI (shading) from three biological replicates. Note that PCNT mRNA moved away from the centrosome upon the harringtonine or puromycin treatment but stayed close to the centrosome upon the emetine treatment, similar to the control. Courtesy of Li-En Jao, University of California, Davis.
Researchers led by Li-En Jao from the University of California, Davis, are studying how cells rapidly expand their centrosomes before division through a process called centrosome maturation. When this process doesn’t work properly, it can lead to genome instability and the uncontrolled cell division seen in cancer. Using Imaris software and the Andor Dragonfly confocal microscope system they have made new discoveries about how centrosomes organize microtubules during cell division.
Centrosomes are micron-scale, self-assembling organelles without membranes. Before cell division, centrosomes “mature” so that their microtubule-organizing activities are at the maximum necessary to support organization of spindle microtubules.
For the new study, the researchers focused on the protein pericentrin, which initiates centrosome maturation by helping to recruit other proteins. They wanted to learn more about how cells can produce and deliver large quantities of this large protein to the centrosome in a matter of minutes.
Detecting weak signals
To learn more about how and where pericentrin is produced in a cell, the researchers combined single molecule RNA in situ hybridization (smFISH) and antibody staining to detect both pericentrin RNA and newly synthesized pericentrin protein. Co-localization of RNA and newly synthesized protein revealed the exact RNA molecules being translated.
The researchers imaged pericentrin RNA and proteins in cultured human cells using the Andor Dragonfly system, a spinning disc confocal microscope that incorporates a microlens confocal scanner with high sensitivity cameras. “The Dragonfly system offered the high sensitivity and confocality to detect the weak smFISH signals,” said Jao. “It also allowed us to image both pericentrin mRNA and protein with high sensitivity and resolution.”
Next, the investigators used Imaris to quantify how pericentrin RNA molecules were distributed in relation to centrosomes at the single cell level. They began by quantifying the 3-D RNA distribution within the cell by using Imaris to convert the protein signal into surfaces and the mRNA signals into spots of different sizes in deconvolved images of each confocal z-stack. They then quantified pericentrin protein intensities at the centrosome by placing the surfaces of the protein pericentrin signals over the original images and using the statistics function in Imaris to obtain the intensity sum of the original images within the fit volume.
Pericentrin is built and transported at once
Overall, the image analysis revealed that pericentrin proteins are co-translationally recruited to centrosomes during mitosis. This means that cells build and transport pericentrin simultaneously by using RNA molecules as templates to build proteins and ribosomes to assemble the building blocks in the correct order.
The researchers plan to continue this line of study by using the Dragonfly system and Imaris to find out how the cell coordinates co-translational targeting of pericentrin to the centrosome with recruitment of other centrosomal proteins during centrosome maturation.
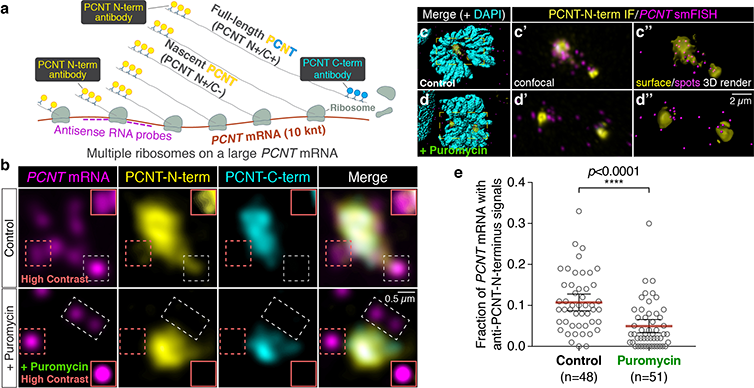
The researchers used smFISH and double immunofluorescence (IF) to distinguish between newly synthesized and full-length pericentrin (PCNT) proteins (a). (b) Prometaphase HeLa cells received PCNT smFISH and anti-PCNT immunostaining against the N- and C-terminus of PCNT protein. Putative active translation sites were labeled by PCNT N-term IF and PCNT smFISH, but not by PCNT C-term IF (top panel). Upon the puromycin treatment, PCNT N-term IF signals were no longer colocalized with PCNT smFISH signals, indicating that those PCNT N-term IF signals on RNA represent nascent PCNT polypeptides. Orange boxes show higher contrast of areas labeled with dashed orange boxes. (c-d) Examples of rendering the PCNT protein and PCNT mRNA signals into “surface” and “spot”, respectively, from a confocal z-stack images using Imaris. (e) PCNT smFISH signals between 1 and 3 µm radius from the centrosome center were quantified for the presence of anti-PCNT N-term IF signals with or without a short puromycin treatment. Courtesy of Li-En Jao, University of California, Davis.
Author: Li-En Jao and colleagues, University of California, Davis
Category: Case Study
