Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Andor's Benchtop Microscope is a highly capable yet compact imaging system. A focus on ease of use has made it possible for new users to start imaging in a much shorter time than other systems. As the system draws upon technology in our high-end Dragonfly confocal systems experienced users can also perform their imaging experiments including 3D volumes and montages, without compromising on image quality or speed. We have now developed a super resolution mode for our benchtop confocal microscopes making it possible to gain further insights into the inner workings of the cell with ease.
Normal light microscopy provides a lateral resolution that is diffraction limited to around 250nm. Unfortunately, this is not enough resolution to resolve many subcellular structures and processes of interest within the cell. Over the years microscopists have developed many ways to push past the diffraction barrier and reveal further information. Techniques such as STORM, STED and DNA-PAINT provide exceptional resolutions and have been very effective in improving our understanding transport pathways, and interactions of different proteins and other fundamental aspects of the cellular machinery. An excellent overview of super resolution techniques is presented in this webinar: What Is Super Resolution Microscopy?. Many variants of super resolution have been developed, each trying to offer more resolution, greater compatibility with live cells, or other practical benefits. Despite these advancements, there remain some limitations to making super resolution accessible to a wider range of users. This includes the need for expensive and specialised imaging systems, dedicated labels with new labelling protocols to learn and validate, and need for high-power densities which are not conducive to live cell imaging. Furthermore, running the super resolution system can be very challenging for less experienced users when confronted with an array of technical parameters to adjust and optimise.
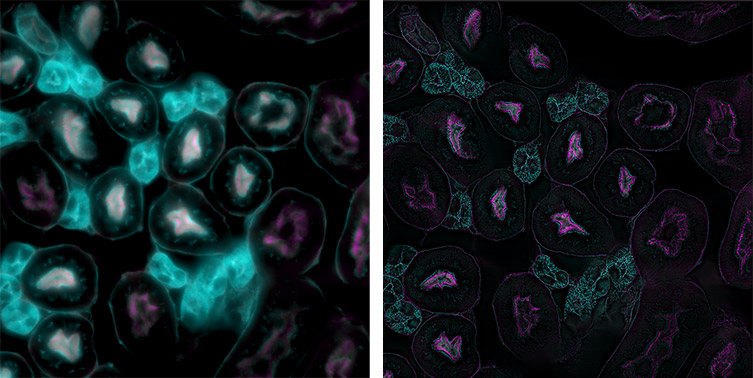
Figure 1: A comparison of widefield and super resolution on Andor's Benchtop Microscope. The normal widefield image is shown on the left and the super resolution mode enabled on the right. Sample: FluoCells™ Prepared Slide #3 (mouse kidney section with Alexa Fluor™ 488 WGA, Alexa Fluor™ 568 Phalloidin, and DAPI), Invitrogen.
“SRRF” (Super Resolution Radial Fluctuations) offers a software-based approach to super resolution that is compatible with normal fluorophores, operates at low illumination intensities on normal microscopes, and is thus much more accessible than many other techniques [1,2]. It can provide a useful boost in resolution that allows subcellular information to be studied. Andor’s SRRF-Stream+ is a further exclusive development of this method that leverages optimised GPU processing to allow SRRF processing to be performed in real time and with improved image quality on compatible cameras [3,4,5]. SRRF-Stream+ has proven popular with many users finding it an effective way to perform super resolution imaging in 2D [6,7,8,9]. However, it is still necessary to understand the SRRF parameters and adjust these iteratively to get the best result for a given specimen [10].
Andor’s Benchtop Microscopes seek to combine high quality imaging, in a compact format, while striving for ease of use throughout imaging experiments. Andor has now brought the same philosophy to super resolution to make super resolution more accessible and easy to use. To do this we have sought to eliminate the need to understand all the technical parameters and settings of super resolution and distil these into sample and workflow related settings users can understand. We have rigorously characterized each input parameter and their interaction on a range of samples to determine optimal settings.
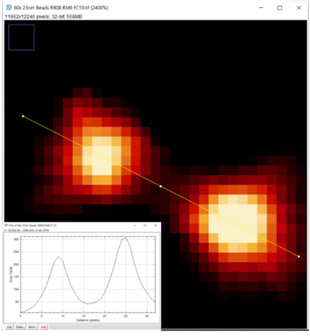
Figure 2: A project to characterise super resolution settings for SRRF-Stream+ was conducted to assess resolution performance, image quality and artefact rejection as well as other factors such as photobleaching and processing time. It was possible to determine the effect of the making small incremental changes to various parameters such as shown here for the ring radius parameter. The selected image shown here is for a 60x 1.42NA objective and fluorescent bead sample. At a ring radius value of 0.8 the normal profile of the fluorescent bead is maintained. Distortion appears as the ring radius is reduced further (data not shown).
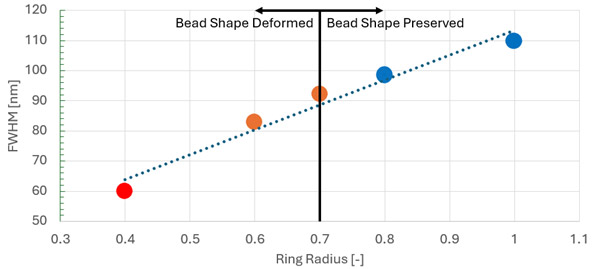
Figure 3: Example data showing analysis of the ring radius parameter on resolution and maintaining integrity of the source data using 23 nm fluorescent beads. Here we can see that for the 60x 1.42NA objective, a value of 0.8 gave optimal resolution (lowest FWHM value) before bead shape profile deformation appeared in the super resolution output. The radiality magnification (RM) and frame count (FC) were fixed at RM=6 and FC=10, respectively, for this study.
Following these experiments to characterise the SRRF-Stream+ parameters for the benchtop microscope we are now able to provide a simple and effective workflow. Settings are configured into 2 easy to understand steps. The first is by sample type, the second step is by processing speed.
Settings for different sample types have been configured into 2 options:
This setting determines the processing speed you require and has 2 options:
Aside from these settings, it is important to set a suitable exposure time to provide a sufficient Signal to Noise and take care preparing the specimen to begin. In other words, the final result will depend on the quality of the input data. As is true for all successful fluorescence imaging, the best image results come from the best samples. Full information on how to set exposure time to achieve suitable signal to noise using the data histogram is provided in the Benchtop Microscope User Guide.
Super resolution on Andor’s Benchtop Microscope finally becomes simple as the complex technical parameters have been eliminated. The workflow can be summarised as shown below:

A full field of view super resolution image can be obtained in as little as 5 seconds and around 15 seconds for “Fast” and “High Quality” modes, respectively. Optimized SRRF-Stream+ resulted in ~2.5 reduced FWHM that transforms in much sharper final image. A final resolution of 140-180nm is anticipated, which will depend on sample type, preparation, and resulting fluorescence signal-to-noise. FWHM measurements are shown below:
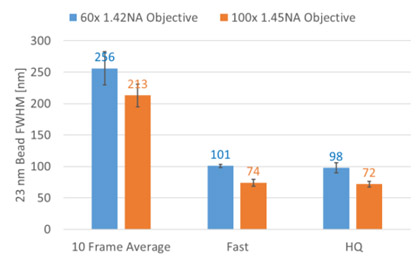
Figure 4: Andor’s Benchtop Microscope (confocal mode) FWHM values (10 frame average) for recommended 60x and 100x objectives vs computed FWHM values for both Fast and High Quality super resolution modes. The FWHM value is approx. 2.5 lower when super resolution Fast or High Quality modes are applied.
SRRF-based methods have proven useful to study cellular structure, co-localisation and other sub-cellular components within the resolution capabilities of the technique [11,12,13,14,15,16]. It is ideal for fixed cells, but it is possible to image live cells by reducing the region of interest to enable a faster frame rate, providing the labelling is relatively bright. For greater resolving power, other techniques such as STORM or DNA-PAINTcan be more applicable which would require the Andor Dragonfly confocal imaging system.
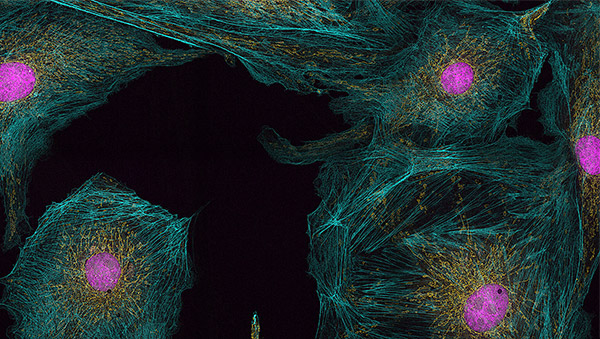
Figure 5: Super resolution image from the Andor Benchtop Microscope - BPAE prepared slide #1 60x, 1.42NA, widefield EPI. Yellow: Mitochondria labelled with MitoTracker™ Red, Blue: F-Actin labelled with CMXRos Alexa Fluor™ 488 Phalloidin, pink: nuclei labelled with DAPI. A high contrast, detailed image of actin and mitochondrial networks is revealed without the image blur of widefield.
Super resolution permits imaging beyond the diffraction limit, enabling additional insights to be obtained for a host of cellular processes making it of great value to many researchers. Many super resolution techniques have evolved to address specific needs in terms of raw resolution or compatibility with living cells. In general, super resolution methodologies may be restrictive in terms of their requirements for specialised equipment, or labelling strategies making them less accessible to many researchers. Following extensive testing of the parameters of the SRRF-Stream+ super resolution method it has been possible to determine combinations of settings that apply to sample type and processing speed. This has been incorporated in a Super Resolution module in Andor’s Benchtop Microscopes. The result is that Andor’s Benchtop Microscope introduces a super resolution capability that offers true ease of use, while providing a resolution enhancement that can be useful for a range of applications.
Find out more at Microscopy Imaging Systems
Date: May 2024
Author: Alan Mullan, Boris Odlozilik, Alexander Stasheuski and Allister Patterson
Category: Technical Article
