Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Using Spinning Disk Confocal Microscopy in Biomedical Research
The Department of Cell and Chemical Biology at the Leiden University Medical Center (CCB-LUMC) aims to unravel the molecular keystones of cellular function and understand their perturbations in disease. The ultimate long-term goal is to translate the emerging knowledge and technologies to clinical practice. To accommodate this mission, research spans from basic understanding of cell biological processes to chemical manipulation of complex cellular systems (such as organoids and tissues), paving the way to intervention in disease. This research stretches across several subjects in life sciences: viral and membrane biology, cell biology of chemotherapeutics, oncogenic signalling, antigen presentation in diabetes, among others.
The method of choice for studying the relevant molecular mechanisms in action is live-cell imaging using confocal microscopy. The Andor Dragonfly spinning disk confocal is widely used for various research projects at the LUMC Cell and Chemical Biology Department. The CCB department uses time lapse microscopy to understand cell biological processes and spatiotemporal behaviour of cells, organelles, and molecules, thus providing considerably more information relative to that obtained using fixed samples.
The use of CRISPR-mediated knock-in technology, which is rapidly becoming standard practice, synergizes with confocal time lapse imaging to allow observation of endogenous fluorescently tagged proteins in living cells. However, since knock-in(s) are under the control of endogenous promotors, their expression levels are usually low, especially when compared to ectopic (over)-expression levels. Therefore, the CRISPR knock-in(s) analysis demands a microscope that delivers extremely sensitive detection.
Furthermore, research on organellar behaviour and studies in mitotic cells encounter additional technical challenges related to iterative acquisition speed and phototoxicity.
"The Andor Dragonfly provides us solutions to the challenges of sensitivity, phototoxicity without compromising acquisition speed."
Dr Lennard Voortman, Department of Cell and Chemical Biology at the Leiden University Medical Center

Figure 1 - Andor Dragonfly 500 at the light microscopy facility of the Leiden University Medical Center
In this section, we present an overview of several research projects from the laboratories of Professor Jacques Neefjes and Professor Huib Ovaa at CCB-LUMC. These projects vary from basic mechanisms of cell biology to translational research, employing modern microscopy and molecular biology techniques using the Dragonfly spinning disk microscope. (For an overview of Dragonfly spinning disk please read Browne M., 2017)
The complexities of organellar movement through cellular space are intimately linked with their functional states. Motile organelles (mitochondria, endosomes, lysosomes and autophagosomes), and infectious agents (including bacteria and viruses) move along microtubules. The movement is done in a bidirectional stop-and-go fashion and is driven by the kinesin motor proteins and the dynein-dynactin motor.
Several biological questions remain unanswered:
To answer these questions the Neefjes laboratory has made several double-color knock-in cells monitoring different potential components of the bidirectional movement pathway. Examples of knock-in(s) include endogenous motors (KIF5 and dynein), or late endosomal effector proteins (RILP and FYCO1). The late endosomal proteins act downstream of the well-known small GTPase Rab7. Another tool used is a cell line is where all vesicles are labelled; the use of this cell line will allow the analysis of the “crowdedness “of the vesicles in cells.
The imaging of this research project will be performed using time-lapse microscopy experiments of living cells followed by extensive image analysis. The final aim of the work is to understand the basis of bidirectional organellar transport in cell biology. (see Figure 2 as an example)
In a similar line of research, chemical inhibitors designed to specifically target the deubiquitylating enzyme USP32 were employed in combination with the cell lines to chemically perturb intracellular transport pathways. The deubiquitylating enzyme USP32 modulates Rab7 function (Sapmaz & Berlin et al, 2019).
In conclusion, using the various chemical biology tools from the Ovaa laboratory, the Neefjes laboratory will continue to dissect the intracellular organelles bidirectional movement mechanism. Extensive live imaging microscopy will be required to push this project forward.
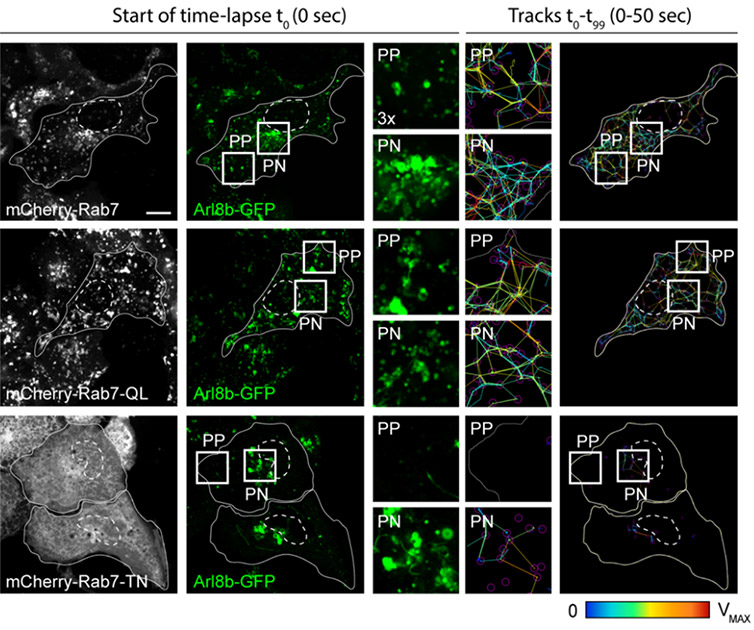
Figure 2 - Representative Dragonfly 500 confocal images of bidirectional movement and organelle dynamics. Representative confocal images of live HeLa cells expressing mCherry-Rab7 or its mutants, together with Arl8b-GFP (green), taken at the start of time-lapse (t0). Right panels: tracks followed by Arl8b-positive vesicles during the time-lapse lasting 50 s recorded at 0.5 s per frame. Time-lapses were collected on a Dragonfly500, and vesicle tracking was performed using TrackMate for Fiji. Image adapted from Jongsma, Bakker et al, EMBO Journal 2020
The endoplasmic reticulum (ER) is the central intracellular compartment that can interact with each and every intracellular organelle. The ER is a complex biosynthetic organelle that also secretes glycol lipids and ions and transduces calcium signalling and metabolic information to other organelles. To compartmentalize and facilitate these diverse functions, the ER harbours a dense perinuclear fraction and a more tubular peripheral network.
The results of this research were published in EMBO Reports (Spits et al., 2021) and present nearly 50 movies acquired using spinning disk confocal microscope, the Dragonfly spinning disk microscope.
Endosomes provide membrane and protein components that are crucial for successful cytokinesis and abscission. The Neefjes laboratory has recently identified an ER-embedded ubiquitin ligase that controls the endolysosomal pathway's perinuclear location (Jongsma & Berlin et al., 2016). The control of this localization is a crucial element for efficient and timely cargo exchange. However, we expect in mitosis, this intercompartmental organization (i.e., the interaction between the ER and endosomes) may be lost to facilitate cytokinesis and distribution of the endolysosomal system to the nascent daughter cells.
To further understand the spatiotemporal organization of endosomes (and other organelles) during mitosis—and the effects of the ER thereupon — genome editing techniques will be used in combination with spinning disk microscopy to follow fluorescent mitotic cells in real-time. The researchers - Ilana Berlin and Lennard Voortman - add that “this enterprise will require extensive microscopy time-lapse imaging, which is a key application for the Dragonfly confocal”.
Date: May 2021
Author: Dr Ilana Berlin, Dr Lennard Voortman & Dr Claudia Florindo
Category: Application Note
