Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Challenge Background
Understanding the molecular basis of development, brain function, neurodegenerative diseases, cancer, and behaviour is an enormous task. Up until recently, RNAs could be sequenced in bulk, or at the single-cell level, but unfortunately, the tissue environment information was lost. With smFISH (1) (single molecule FISH), the spatial information is retained, but this is restricted to a small number of genes that can be spectrally separated.
Gathering the 2D tissue environment information and (if possible, 3D) is essential to establish the network of gene expression which underlies tissue function and disease. The Holy Grail of modern cell & molecular biology, is therefore, to probe the gene expression network built with information from mapping multiple (Xn) RNAs in the same sample.
Multiplexing in cell biology is the detection of numerous (Xn) RNAs (or other biological molecules) in a tissue /cell, either in its 2D or 3D context. Multiplexing is becoming a very hot topic in neurosciences, oncobiology, disease target diagnostics, development and behavioural studies among others. Due to its power in providing the spatial and sequence information of multiple gene products, the number of applications in which new-era technology will undoubtedly make a difference is expected to continue to increase rapidly. It is important to remember that these technologies can appear under many names in the literature: spatially resolved transcriptomics, spatial transcriptomics, spatial genomics, and in-situ multiplex imaging.
In RNA biology, several techniques have been developed to allow in-situ multiplexing. Each has its advantages and disadvantages, but all have the same goal of building a spatial (visual) map of gene expression and connectivity to its cellular and tissue context. Examples of such techniques are FISSEQ (2), instaSEQ (3), osmFISH (4), STARmap (5), MERFISH( 6) and seqFISH (7). The relevance of these methods is growing in the scientific community. An example of its importance is that spatially resolved transcriptomics was elected 2020´s method of the year (by Nature methods).
To identify the expression of genes within tissue, several rounds of hybridisation and detection need to be performed sequentially in the same sample. The sequential treatments and image acquisitions are performed using a microfluidic system attached to a microscope. Fluorescent probes label the hybridised RNA molecules, the image data is acquired (typically a volumetric montage is scanned), and the probes are washed away. After each image dataset is acquired, a “strip and wash” step is followed by another hybridisation round. This procedure will be repeated N times and results in large volumes of encoded image data. An example of one such dataset is shown in Figure 1, and results from a six round experiment using the STARmap protocol. Each spot in each volume is coded by colour and represents one nucleotide in the RNA sequence. Following, the multiplex labelling and data acquisition steps, an off-line analysis pipeline is used to translate this data into a map of gene expression of all the genes targeted for a specific experiment.1
Several challenges need to be overcome to achieve proper multiplexing experiments. Fundamental technological topics include automation procedures, speed of acquisition, sensitivity and resolution, photobleaching, uniformity and throughput. Once the data has been acquired, there are several bioinformatics challenges to be considered. The data from each field or volume must be spatially matched, even in the presence of shifts and non-linear shape changes, so that the sequentially probed nucleotides (ACUG) can be sequenced and unequivocally assigned to a gene product or mRNA molecule. Consistent and reliable data needs error tolerance or error checking.
Technology Solution
A camera-based multipoint confocal system is an excellent solution for multiplexed in situ imaging. Any such system is required to provide very efficient background rejection as well as high sensitivity and resolution to deliver accurate localisation of the target gene products. Due to the complexity and expected duration of the experimental procedures, speed of acquisition is a crucial parameter for high throughput and productivity. Hence, the ability to acquire multiple wavelengths simultaneously is a desirable feature.
Since the same volume of the specimen must be repeatedly scanned or imaged in each round of the procedure, high stability is also critical. The system must retain focus over many rounds of imaging and sequential labelling during which the temperature and conditions within the flow cell undergo substantial changes.
The acquisition system should be optimised for gentle imaging, delivering data with minimal photobleaching. To achieve this the use of high sensitivity, high dynamic range detectors is vital, not only to allow low light acquisition, but also to capture the full range of signals in the sample. These may vary over several orders of magnitude, especially when the nuclear DNA signal is captured as part of the procedure.
When capturing high resolution volumes which extend over millimetre squared areas and deep into the tissue, the ideal imaging system should offer highly uniform illumination across the field of view for all wavelengths used. Montage data sets acquired with high uniformity provide the best quality input data to the analysis pipeline, resulting is consistent SNR across volumes and aiding accurate detection, identification and mapping throughout the tissue volume.
Spatial transcriptomics can take several hours per round to execute, so that 6-10 round protocols can last for more than 24 hours. It is essential therefore that the, the software, hardware and microfluidics systems can be integrated and synchronized in their various tasks. Ideally, this integration should allow flexible and customisable protocols that can be adjusted to experimental requirements.
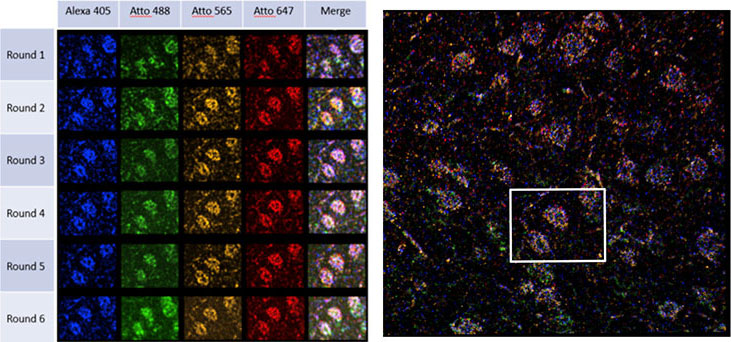
Figure 1 – Mouse hippocampus sequential labelling by STARmap, imaged with Dragonfly. Left image shows MIP of one field of a montage with 4 channels and 71-Z sections over a depth of 20 um; right image shows a MIP of the montaged data set. Data courtesy of Dr Abbas Rizvi, Columbia University, Genomics Center, New York.
Andor Solutions for Spatial Transcriptomics Studies
Dragonfly is an excellent solution for in-situ multiplexing hybridisation. As a multipoint confocal, Dragonfly will deliver high-quality confocal images at high speed, (up to 400 fps) in confocal mode. Equally important in tissue scanning is the large field of view (FN 22) which allows for rapid acquisition of large tissue volumes. Additionally, the dual microlens disk system combined with Andor´s high dynamic range and high QE cameras such as back-illuminated Sona or iXon EMCCD series allows the capture of all the signals in the sample: from the dimmest to the brightest.
When acquiring data for multiplexing in-situ experiments, there is a substantial requirement for uniformity and high throughput. Using Andor´s patented Borealis illumination system, Dragonfly delivers excellent uniformity across the full field of view; further, by acquiring multiple fields of view as well as imaging different wavelengths simultaneously, the throughput increases considerably.
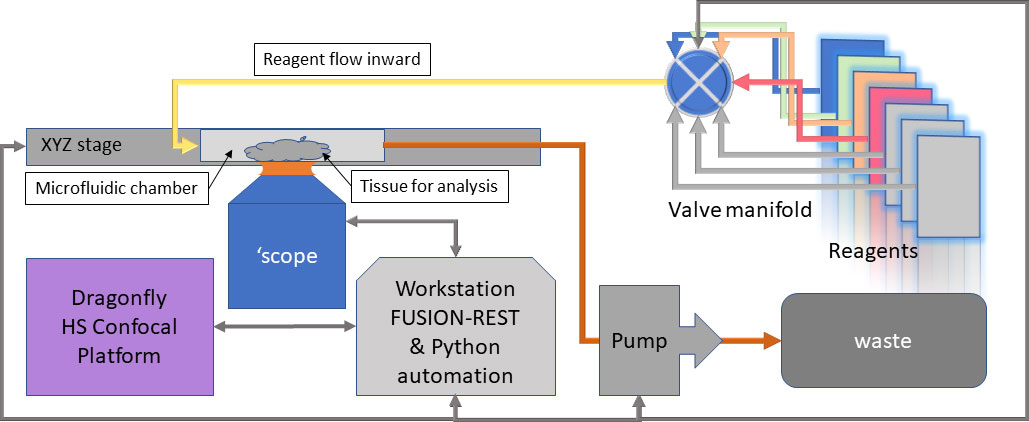
Figure 2 – Block diagram of Dragonfly-based system with microfluidics for In-Situ Sequencing (ISS)
The recently incorporated REST-API built into Fusion software, allows the Dragonfly to be controlled from an external program e.g. Python, LabView, MatLab. This provides the capability to communicate with the microfluidics system for sequential reagent flows for hybridization, labelling and washes, while Dragonfly manages the imaging hardware and can be triggered on demand to acquire spatially consistent, multi-channel volumetric data. Changes to the fluidic and imaging protocols are therefore easily controlled by the user, once familiarity with the coding environment is achieved. A block diagram of such a system is shown in Figure 2.
Dragonfly’s unique blend of high speed, high resolution imaging combined with large field of view, excellent background rejection and Borealis illumination system provides a powerful tool for in-situ sequencing (transcriptomics). Extending these capabilities with the REST API provides researchers with the tools needed to create flexible, +highly multiplexed imaging platforms,
| Key Requirement | Multiplexing in Situ Experiments: Dragonfly |
| Fast acquisition speeds | Dragonfly is at least ten times faster than point scanner confocal systems. Two camera ports enable Dragonfly to acquire two channels simultaneously. EMCCD & sCMOS detectors allow very low light imaging and high acquisition speeds. Result 1 – Increase productivity by acquiring confocal images faster. Result 2 – Increase productivity by detecting two independent channels simultaneously. |
| Uniform illumination | Uneven illumination results in images where signal intensity varies for the same molecule in different locations: if uncorrected, quantification of RNA molecules would be inaccurate.Andor´s patented Borealis illumination system delivers uniform illumination across the field of view. Result 1 - Acquired images deliver accurate and quantifiable data. Result 2 - Multiple fields of view can be stitched to deliver consistently uniform spatial maps. |
| Automation control | Intercommunication of the different instruments required to perform a multiplexed experiment is essential. Fusion’s REST-API allows Dragonfly to be easily triggered for imaging and synchronized with other devices for multiplexing. Result 1- Automatic control of the multiplexing set-up. Result 2 - Flexible protocols that can be adjusted during the experiment. |
| High sensitivity 3D tissue imaging | High QE and low read noise combined with low background and high signal throughput in Dragonfly, provide an excellent combination of features for this application in native or cleared tissues. sCMOS cameras are 3 to 5 times more sensitive than point scanning detectors, with up to 95% QE, and a high dynamic range (16 bits – 65 535 grey levels). Result 1 – Acquire all the signals in the image, avoid saturation and ensure proper representation of cellular RNAs. Result 2 - Use shorter exposure times for acquisition. Increase speed of acquisition and productivity. Result 3 - Use low laser power for imaging. Decrease photobleaching. |
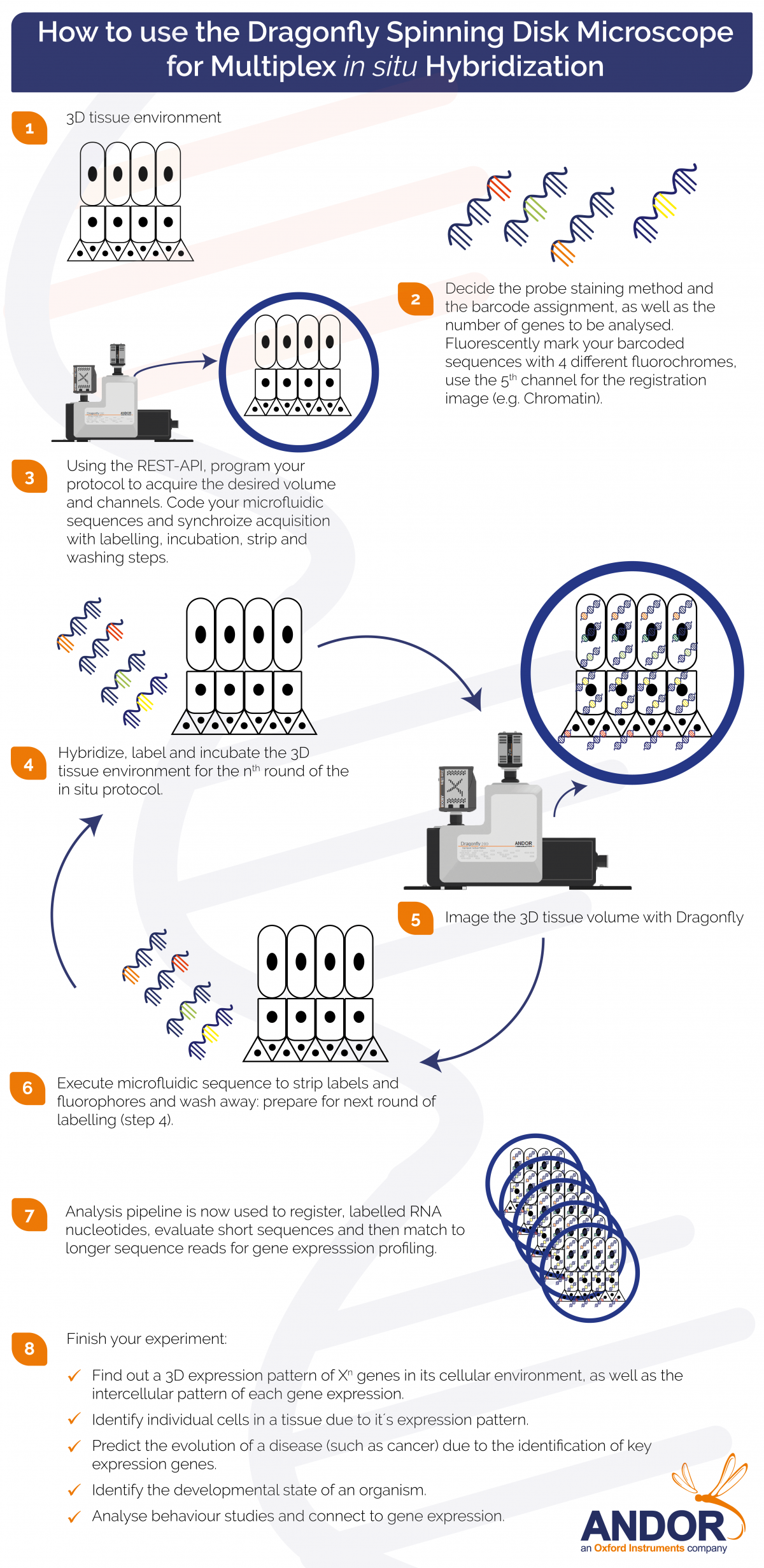
Please take a look at our infographic to visualise the required steps in Spatially Resolved Transcriptomics Experiments (in-situ multiplex imaging) experiments.
1. Multiplexing increases identifiable gene products (GP) rapidly. With 4 labels per round, R the number of GP’s which can be labelled uniquely is 4R. Theoretically, nine rounds could label all gene products in the human genome!
Date: Aug 2020
Author: Dr Claudia Florindo and Dr Mark Browne
Category: Solution Note
