Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Challenge Background
Viruses pose a significant threat to human health; they are responsible for a wide range of diseases and are found throughout the environment. Understanding how viruses cause infections, virus-host interactions and adaptability are crucial to developing effective anti-viral treatments. Light-microscopy methods can be incredibly useful for imaging viruses and related cellular processes, which can lead to a better understanding of viral infections and management strategies.
Imaging of viruses poses several experimental challenges, being one of the critical challenges their small size. Typically, individual virions, or virus particles, are less than 220 nanometres (nm) in diameter, which is at the diffraction limit of light. Advanced imaging methods are therefore required to bypass this limitation. Using advanced imaging methods (e.g. super-resolution microscopy) objects like viruses, that are smaller than 220 nm, can be accurately resolved. Nevertheless, when imaging active viruses, other demanding challenges such as the difficulty to label viral envelop components, phototoxicity to the host cell, photobleaching, also need to be addressed. Last, but not least, the viral infection process can occur over the span of minutes which requires the acquisition of multiple images (high temporal resolution) to capture all the events of the infection.
Technology Solution
Spinning-disk confocal microscopy is an ideal solution for imaging virions as well as capturing live-cell virus-host infection events.
Spinning disk confocal technology delivers gentle (live-cell compatible) high-resolution images without compromising cellular viability.
Speed and Sensitivity – Due to the timescale of infection events (several minutes), a high number of images need to be collected to capture all the infection steps. The signal from the labelled virus will be low, but photobleaching of the sample or phototoxicity to host cells must be avoided. A camera-based detector is therefore also required to provide the necessary sensitivity at high speeds.
Resolution – The resolution required to image viruses l is in the XY range of 50-200 nm. The diffraction limit of light microscopes is around 220 nm; therefore, techniques that can overcome the diffraction limit, i.e. super-resolution techniques are required.
Live Cell Optimised Super-Resolution – Super-resolution techniques that overcome the diffraction limit of the light microscope generally require the acquisition of a vast number of frames (on the order of the 1,000 to 10,000 images) and/or imaging with extremely high light intensities. In most cases, sample preparation is also complicated, and there is also the requirement of specific fluorophores. These requirements render most currently available super-resolution techniques incompatible with live-cell imaging. Additionally, the use of specific super-resolution fluorophores may alter the normal activity of host cells or viruses.
Andor Solutions for Imaging Viruses
The Andor Dragonfly Spinning-Disk Confocal platform is the complete and ideal solution to study viruses due to the perfect combination of speed, sensitivity and resolution. Imaging of viral envelope components is possible with Dragonfly coupled with Andor´s high sensitivity cameras, such as the back-illuminated Sona sCMOS or iXon EMCCD series. Dragonfly allows imaging modalities such as confocal spinning disk, TIRF, and dSTORM. All these imaging modalities are delivered in one single system, allowing the user to select the best match for each specific application.
A further benefit is that any imaging modality can be combined with the live-cell compatible super-resolution technique “SRRF” (super-resolution radial fluctuations). Unlike other super-resolution techniques, SRRF is compatible with live cell, offering the advantage of high-speed super-resolved live imaging. SRRF-stream (Andor’s GPU-based, optimised implementation of SRRF) is available on the Dragonfly and allows the acquisition of super-resolved images in an instant. Additionally, SRRF-Stream is universally compatible with the use of any fluorophore used to label viruses.
And finally, Imaris provides a robust package of analysis solutions adapted to all the fields of life sciences including 3D analysis, visualisation, segmentation and rendering tools. Able to handle large data sets, Imaris is ideal for analysing long-term, virology imaging studies.
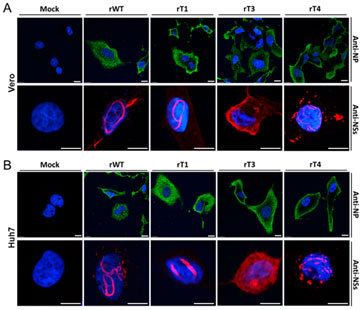
Figure 1 - Analysis of filament formation in virus infected cells. Figure presents images of fixed Vero (A) and Huh7 (B) cells infected with RVF virus. Images were obtained using Andor Dragonfly. The infection and filament formation was analysed by immunofluorescence microscopy using anti-NP (in green) or anti-NSs antibodies (in red). Nuclei were stained with DAPI (in blue). Scale bars=10 μm; NP=nucleocapsid protein; NS=key virulence factor of RVF virus. (reference: Shufen, L. et al. ,2019 - Viruses.)
| Key Requirement | Virus Imaging Solution: Andor Dragonfly and High QE Cameras |
| Speed and Sensitivity | Dragonfly is at least ten times faster than point scanning confocal systems. The internal beam splitters allow Dragonfly to acquire two channels simultaneously on two independent high sensitivity sCMOS or EMCCD cameras. Result 1 – Detect fast events with acquisition speeds up to 400 frames per second in confocal mode. Result 2 – Detect ultra-fast events with acquisition speeds up to 1500 frames per second in Widefield or TIRF mode. Result 3 – Detect two independent channels simultaneously without compromising speed or resolution. |
| Multi-Modal Imaging Platform | Andor Dragonfly is truly a multi-modal imaging platform. Dragonfly accommodates the following imaging modalities: 1) Spinning-Disk Confocal, 2) Super Resolution dSTORM, 3) TIRF, and 4) Laser Widefield. Result 1 – Use the best imaging modality for or application (no need to have multiple imaging systems). Result 2 – Use different imaging modalities in the same sample (e.g. confocal and TIRF) - extract more pieces of information from a single sample. |
| Extended Spectral Range Use NIR wavelength | The Borealis illumination system, together with the multimode optical fiber delivers very flat illumination across all the excitation wavelengths from the UV to the NIR (400-800 nm). Result 1 – Use multiple probes (400-800 nm) to capture information of different cellular structures simultaneously. Result 2 – Perfect stitching and accurate quantification of your images are delivered due to the high uniformity of the Borealis illumination system. Result 3 – Image live host-cell virus interactions using the low energetic radiation of the NIR wavelengths. Result 4 – Increase Signal/Noise ration of your data by acquiring images using NIR wavelengths. (Samples present very low autofluorescence in the NIR range). Result 5 – Capture infection events deep inside cell and tissues due to the high penetration of NIR wavelengths. |
| Super-resolution | Dragonfly supports dSTORM and TIRF. Using the motorised illumination zoom, Dragonfly delivers high-density power required for single-molecule localisation microscopy. Simultaneous dual-colour TIRF is possible with the same sample penetration for both wavelengths. Result 1 – Acquire very detailed information of virus, with high spatial resolution. Images can have up to up to 20 nm XY resolution using dSTORM. Result 2 – Acquire 3D high-resolution structural data. Dragonfly delivers single-molecule 3D dSTORM data due to the use of an astigmatic lens. Result 3- Image live virus-membrane infection events using dual colour simultaneous TIRF imaging with axial resolutions up to 50 nm. |
| Live-Cell Compatible Super-resolution (SRRF-Stream) | SRRF is compatible with conventional fluorophores, and there is no need for complex sample preparations. SRRF is compatible with confocal, TIRF and widefield imaging, and with deep imaging. Result 1 – SRRF-stream boosts resolution by 2- to 6- fold (50 - 150 nm resolution) in the final data. Result 2 – Due to its low power requirements (mW/cm2 to W/cm2 range), SRRF-stream is compatible with live-cell imaging. Result 3 – The SRRF-stream algorithm allows acquisition of live cell images that break the diffraction limit at a frame rate of 10 frames per second. |
| Complete Analysis Solution | Imaris software provides a full package of analysis solutions adapted to all the field of life sciences, including virology. Imaris for tracking provides tools for automatically analysing moving objects, create a lineage tree plot and generate quantitative information from your live image data. Result 1 – Quantify, analyse and 3D render data acquired with the Dragonfly and Andor’s cameras. Result 2 – High-quality movies can be created with Imaris analysis software. |
Date: June 2020
Author: Dr Kalpana Iyengar & Dr Claudia Florindo
Category: Solution Note
