Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
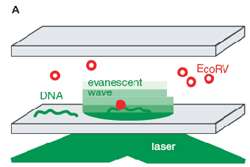
Figure 1 – Single molecule TIRF microscopy setup for the study of DNA – EcoRV enzyme interactions.
Structure-function relationships in DNA offer an unlimited source of fascinating scientific challenges, dating back to the first quantitative studies of the 1950s. Since the seminal paper by Watson and Crick, several generations of researchers have tackled numerous questions regarding mechanistic aspects of DNA, aimed at better explaining its primary function, that of conservation and propagation of genetic information.
The group led by Dr. Pierre Desbiolles at Laboratoire Kastler Brossel, CNRS and Université Pierre et Marie Curie, Paris, addressed several important questions regarding the way in which certain proteins act upon and modify DNA, to make it either more, or less, accessible.
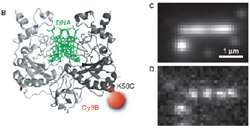
Figure 2 – (B) Cartoon of DNA engulfed by one molecule of EcoRV enzyme labelled with Cy3. (C,D) EMCCD camera images of single EcoRV molecules attached to DNA strand. Image in (C) is composite of several hundred images of enzyme molecules whereas (D) is an individual exemplary frame from composite shown above. Note the inherently low light nature of single molecule image in D.
For the DNA readout process to operate with high fidelity, a plethora of types of proteins must be able to bind onto a DNA molecule to carbon-copy information contained within. Sometimes this information becomes corrupted, or is not readily accessible, for example due to a viral infection or chemical damage to the DNA. If this occurs, a special class of repair / scavenger enzyme proteins remove unreadable parts and continue proof-reading to ensure the integrity of information in the living cell. Our knowledge of the mechanism of such DNA damage, and more importantly repair, can be vital for understanding certain genetic diseases and disorders. One way of dissecting and understanding these processes is to use isolated DNA in so-called 'in vitro' conditions, to test how both DNA and its binding partners behave in a controlled and simplified environment.
Pierre′s team, including Isabelle Bonnet and Andreas Biebricher, used an experimental system whereby individual enzyme molecules can be visualised with fluorescent Cy3 labels attached to an enzyme′s surface. DNA molecules were first attached to a surface, and then enzymes were added to allow for binding and visualisation. Complexes of DNA with the enzyme – in this case EcoRV – were placed in an evanescent field, created by a laser beam bouncing off the glass-water interface, so that the enzymes could be visualized by total internal reflection fluorescent microscopy (TIRFM) (Figure 1).
Cy3 labels attached to individual EcoRV enzymes emitted very dim light, detected by an Andor iXon 860 EMCCD camera, capable of detecting down to single photons originating from Cy3 molecules. Conditions were particularly challenging since each EcoRV enzyme was tagged with only one or two Cy3 labels, coupled with the need to image dynamically (Figure 2).
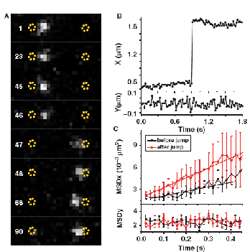
Figure 3 – (A) Jumps of EcoRV along a DNA molecule. White dot represents single enzyme molecule rapidly moving from one end of DNA to another between consecutive frames of rapid low-light video sequence. These jumps were measured to be often in excess of 1 μm (B) which is several hundred-fold more than with initially observed sliding movement. (C) Mean square displacement (MSD) shown in (C) gives additional information on specificity of binding and quality of DNA-EcoRV interaction.
By acquiring extended time series of individual DNA binding events, Andreas and Isabelle confirmed an earlier suggestion that EcoRV could slide along a DNA strand in order to access areas nearby. Such facilitated diffusion has been reported before, but it was unclear whether or not EcoRV could also jump along DNA between fragments which are spaced further apart. The answer to this question was that indeed, long-distance excursions of enzyme on DNA were possible, confirmed for the first time with this direct microscopic observation (Figure 3).
Analyses of several video sequences allowed insight into mechanisms that govern protein–DNA interactions, in particular into the possible regulatory role of proteins that scan often large sections of DNA and readily jump between them. To test their system further, the Desbiolles group performed a series of tests which were designed to show whether different chemical compositions of the experimental environment could affect enzyme jumps on DNA. Numerous salts and buffers were tested in their TIRF setup and subsequent low-light rapid video analysis revealed that length and specificity of jumps were strongly dependent on ions interacting with DNA.
Such a direct single molecule approach resulted in the demonstration that enzyme jumping could accompany previously observed sliding along a DNA molecule. By using TIRF dynamic recordings, it became possible to quantitatively estimate these two processes. The ratio of jumping to sliding, may well differ from protein to protein, depending on both the structure and on the biological function of the protein.
