Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
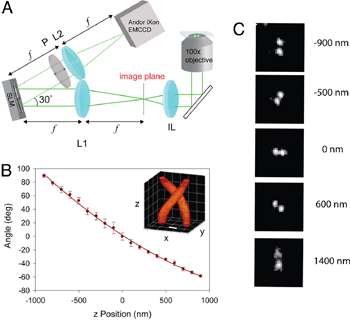
Figure 1: DH-PSF imaging system. (A) Optical path of the DH-PSF setup including spatial light modulator and an Andor iXon3 897 EMCCD. (B) Calibration curve of DH-PSF, (C) Images of a single fluorescent bead used for axial calibration (reprinted from Ref. 1, used by permission)
In the past few years we have witnessed an unprecedented evolution of imaging techniques, directed at helping researchers break through what was previously regarded as an immutable optical resolution limit. Several novel super-resolution methods have made it possible to look beyond ~200 nm into the realm of true nanoscale environments. These breakthroughs have been fueled by the exponential growth of biophysical studies that often called for improved methods, required for precise localization and tracking of single labelled molecules of interest. As such, use of several cutting-edge single molecule fluorescent imaging techniques has made it possible to expand our insights into previously inaccessible nanoscale intracellular structures and interactions.
One such novel tool has been described in a recent paper published by researchers of W.E. Moerner′s group in collaboration with R. Piestun′s group at the University of Colorado.1 M. Thompson, S.R.P. Pavani and their colleagues have shown that it was possible to use a uniquely shaped point-spread function (PSF) to improve image resolution well beyond the diffraction limit in z as well as in x and y.
What makes this PSF different from a standard hourglass-shaped PSF are its two lobes whose 3D projection closely resembles an intertwined helix, lending it the distinctive name of ‘Double-Helix PSF’ (DH-PSF; Fig 1B). The DH-PSF is an unusual optical field which can be made from a superposition of Gauss-Laguerre modes. In the implementation (Fig 1A), the DH-PSF does not itself illuminate the sample.Rather, a single emitting molecule emits a pattern corresponding to the standard PSF, and the standard image of the molecule is convolved with the DH-PSF using Fourier optics and a reflective phase mask outside the microscope. Interestingly, thanks to its shape, the DH-PSF approach can yield distinct images of a fluorophore molecule depending on its exact z position. At the detector, each molecule appears as two spots, rather than one, due to the effectiveDH-PSF response.The orientation of the pair can then be used to decode the depth of a molecule and ultimately helps determine its three-dimensional location inside the specimen (Fig 1C).
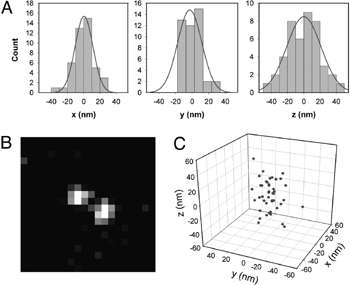
Figure 2: 3D localisation of single molecule. (A) Histograms of precision of localisation in x-y-z. (B) Image of a single DCDHF-P molecule taken with DH-PSF. (C) 3D plot of molecule′s localisations (reprinted from Ref. 1, used by permission)
The usefulness of the DH-PSF has been validated in a 3D localisation experiment involving imaging of a single molecule of the new fluorogen, DCDHF-V-PF4-azide, after activation of its fluorescence. This particular fluorophore typically emits a large number of photons before it bleaches, it is easily excited with low amounts of blue light and it emits in the yellow part of the spectrum (~580 nm), which overlaps well with the most sensitive region of silicon detectors. All imaging has been done with a highly sensitive Andor iXon3 EMCCD camera, operating at 2 Hz and the EM gain setting of x250 (sufficient to effectively eliminate the read noise detection limit). By acquiring 42 images of a single molecule of this fluorophore (Fig. 2B) it became possible to determine its x-y-z position with 12-20 nm precision depending on dimension of interest (Fig. 2AC).
Interestingly, this localisation method allowed the researchers to achieve the same levels of accuracy as those typically obtained with other 3D super-resolution approaches including astigmatic and multi-plane techniques. In addition to this, the DH-PSF method extended the depth-of-field to ~2 μm in comparison to ~1 μm offered by either previously used technique.
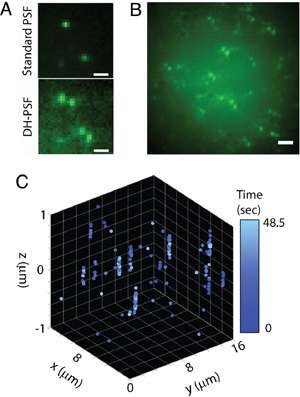
Figure 3: 3D localisation of several DCDHF-P molecules in a thick sample. (A) Comparison between images obtained with standard PSF and SH-PSF (B) Ensemble of several DCDHF-P molecules in 3D space (C) 4D plot of single molecules′ localisations in time during acquisition sequence. (reprinted from Ref. 1, used by permission)
This feature of DH-PSF is particularly useful for imaging of thicker samples that are typically used in fluorescent imaging. Some super-resolution techniques may require samples to be sufficiently thin and adherent to be imaged in a TIRF field for best localisation results. This, however, may prove problematic with some cell types, when membrane ruffling and uniform adherence render TIRF imaging impossible.
The increased depth-of-field obtained with DH-PSF can be noticed in Fig 3A, where we see a comparison between a standard PSF and the helical PSF. One can register individual molecules of another fluorophore, DCDHF-P, with both PSFs, however, the DH-PSF seems to produce images with higher background than the standard PSF. This is partly caused by the helicity of PSF and the presence of its side lobes penetrating a considerable range in the z dimension (see the helix in Fig. 1B inset). What truly matters is the ability of the DH-PSF to achieve particular precision values with equal numbers of photons, and this has been carefully measured in a subsequent study. The method carries the distinct advantage of being able to reveal the molecules′ positions while maintaining approximately uniform intensities throughout the depth-of-field. An entire field of view with tens of individual molecules can be seen in Fig. 3B. The angles represented by such "pairs" are then used to estimate the axial position of a molecule of interest (Fig. 3C).
The Moerner group has further tested their model using higher concentrations of photoactivatable fluorophores in the sample as required for PALM imaging. Similar to previous tests, fluorophore molecules have been embedded in 2 μm thick, synthetic acrylic resin, then repetitively activated, imaged, and localised using DH-PSF.
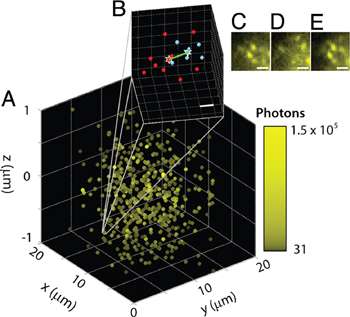
Figure 4: Super-resolved image of high concentration of fluorophore in a thick sample (A). Zoomed in region with calculated 14-26 nm separation in x-y-z (B).(C-E) Activation cycle demonstrating bleaching and subsequent activation of various molecules. (reprinted from Ref. 1, used by permission)
This experiment has confirmed the super-resolving capability of the DH-PSF approach and shown that it was possible to localise and distinguish molecules that are 10-20 nm apart in all three dimensions.
This method, described fully in the original PNAS publication,1 is a notable addition to an expanding toolbox of 3D super-resolution methods. In comparison to multiplane and astigmatic approaches to three-dimensional super-resolved imaging, DH-PSF offers significantly extended depth-of-field. Such a feature makes it possible to “scan” the z-dimension, unravelling precise axial positions of individual molecules within an extended 2 µm sliver of a sample. It is possible that by using improved estimators for DH-PSF this method may become an even more robust imaging tool, allowing for further refinement in accuracy of x-y-z localisation as well as background reduction and increased S/N ratio.
Andor Solutions for Super Resolution
For this particular study the researcher chose to use an EMCCD for its high resolution, speed and sensitivity. The iXon Ultra 897 works well for Super resolution techniques where the electron multiplication can be utilised for weakly fluorescent samples and the fast frame rates with the "Optical Centred Crop Sensor” mode allow the capture of dynamic events. Facilitated by a fundamental redesign, the new iXon Ultra platform takes the popular back-illuminated 512 x 512 frame transfer sensor and overclocks readout to 17 MHz, pushing speed performance to an outstanding 56 fps (full frame), whilst maintaining quantitative stability throughout. The significant speed boost offered in the iXon Ultra 897 facilitates a new level of temporal resolution to be attained, ideal for speed challenged applications such as live super resolution techniques.
