Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Mitotic cell division relies upon the ability of cells to properly distribute sister chromatids into forming cells. To reliably perform the function cells use internal editing mechanisms to correct inaccurate chromosome / spindle fiber attachments. Paired kinetochores are usually aligned to properly attach spindle fibers and segregate chromatin, however, erroneous and misaligned kinetochores do result. Cells contain specialized editing mechanisms to prevent and correct these misaligned kinetochore pairs, although the exact mechanism is not well understood 1,2 . It is postulated by Tatiana Moutinho-Santos at the Universidade do Porto, Portugal's Instituto de Biologia Molecular e Celular (IBMC) that the presence of POLO kinase is necessary to promote chromosome bi-orientation (termed amphitelic arrangement) and thereby preserve the proper kinetochore alignment.
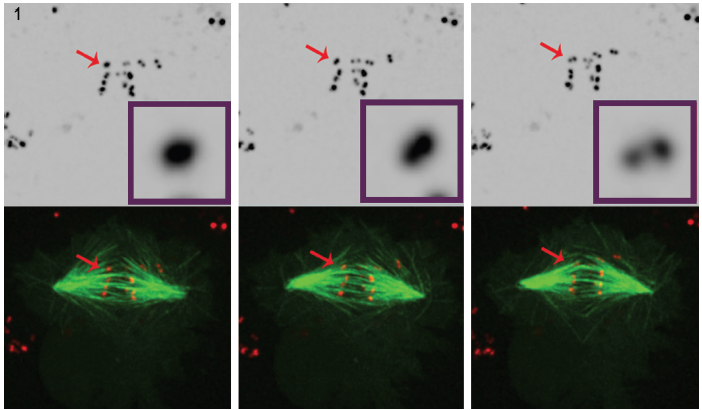
Figure 1. Three Selected still images from a movie of POLO-depleted cells clearly show that, over time, intense centromere/kinetochore signal (red) resolves into a pair of dots, which demonstrates that chromosomes are syntelically attached to spindle microtubules (green). 4D fluorescence microscopy datasets were collected every 30 seconds with 0.5 mm z-steps covering the entire cell volume using a 100°, 1.4 NA plan-apochromatic objective at 25°C with Andor's Revolution spinning disk confocal unit and iXon EMCCD camera, both driven by Andor IQ live-cell imaging software.
To better understand its role in regulating kinetochore development, Dr. Moutinho dos Santos studied depleted Drosophila POLO kinase within live cells to substantiate POLO's involvement in the editing functions required to maintain amphitelic kinetochore arrangement. In this case, syntelic arrangement (sister kinetochores attached to microtubules emanating from the same spindle) was studied. Timelapse analysis of mitosis was performed on Drosophila S2 cells stably expressing CID-mCherry for visualisation of the centromeres, and GFP-a-tubulin for the microtubules. Visualization of kinetochores is demanding due to their small size, low fluorescent signal and brief appearance during cell division. Confocal fluorescence microscopy is often used to image kinetochores, however, even in this environment, visualization remains difficult. The kinetochores are very small (300 nm)3, close to the lateral resolving capabilities of the light microscope, and exceed the microscope's axial resolving ability. Moreover, imaging fluorescent markers within living cells poses potential phototoxic and photobleaching effects. Intense laser illumination can create phototoxicity issues, especially damaging for living, dividing cells.
Andor's Revolution XD spinning disk confocal microscopy is employed to overcome Dr. Moutinho dos Santo's unique challenge associated with the observation of kinetochores. In this case, four separate factors must be simultaneously addressed to adequately visualize the dynamic cellular processes:
These themes of spatial, temporal and intensity resolution recur frequently with fluorescence microscopy and are often at odds with experiments involving the observation of cell viability. For example, long camera exposures and extended periods of high intensity illumination generate the phototoxic effects that damage or destroy living cells. Simultaneously, lightly labeled microstructures demand longer exposure to visualize but can be adversely affected by photobleaching. Compounding the issue is the need to combine traditional three-dimensional data with that of time. Finally, it remains necessary to discern signal from background noise and outof- focus haze.
Speed of acquisition is of particular importance to Dr. Mountinho dos Santos. Control S2 cells exhibit a division cycle of approximately 30 minutes. However, the POLO depleted S2 cells utilized in her experiments showed an arrested period in excess of eight hours. Recording the POLO depleted cell division requires collection of between 7,200 and 12,000 image sets (two fluorochromes imaged every thirty seconds for one to five hours, acquired at 0.5 micron axial steps at up to 20 steps for axial kinetochore resolution). Conventional fluorescence microscopy and laser illumination will permit neither the gentle illumination requirements necessary nor the speed of acquisition to complete this time-sensitive task. For example laser rastering in traditional confocal microscopy requires longer time periods time to collect signal from the sample. Spinning disk confocal systems are used to overcome these traditional barriers and to reveal new insights into molecular kinetochore editing techniques.
Compared to conventional widefield fluorescence systems, the spatial resolution of a spinning disk confocal is superior in both lateral (x and y) and axial (z) dimensions. Through the constant scanning of the pinhole array, samples can be viewed in real-time at high contrast, providing clear images at the diffraction limits of the microscope's optics. This enables the 3D visualization and understanding of the dynamic kinetochore behavior in relation to the microtubules of the spindle. Kinetochore and centromere lateral resolution of 300nm and spindle fiber resolution of 300-500 nm were reported and are detailed in Figure A.
Intensity Resolution
Visualizing the fluorescently labeled kinetochores associated with each centromere places additional demands on the acquisition system. The centromeres and kinetochores under study are primarily visible only during cell division interphase. As the cells pass through prophase, the centromeres have already resolved themselves into Drosophila’s typical twelve chromosome pairs. Managing the available light budget during lengthy acquisition necessitates gentle illumination and high resolution capable by spinning disk techniques. The apertures within the spinning disk unit provide these benefits. Because light excites fluorophores only when an aperture is present, phototoxic effects are minimized. While counterintuitive, a reduced light budget does not indicate faint, hard-to-detect objects. Intensity resolution is increased through the spinning disk's exclusion of out of focus signal and subsequent signal amplification by an EMCCD camera architecture. This creates the new possibility to generate higher contrast images revealing the development and alignment of punctate objects such as kinetochores.
Conclusions
The use of spinning disk technology in Dr. Mountinho dos Santos live cell application led to an important observation. In the absence of the POLO kinase, Drosophila cells were shown to lack the corrective mechanisms necessary to maintain amphitelic chromosome arrangement. The presence of POLO was shown to provide the right environment for correct centromere architecture, while simultaneously ensuring proper chromosome bi-orientation. Cultured Drosophila cells undergoing mitosis in the absence of POLO kinase chromosomes attach to spindle fibers with syntelic orientation, i.e., with sister kinetochores attached to microtubules that come from a single spindle pole.
This conclusion was drawn by careful analysis of 4D fluorescence microscopy in cells expressing fluorescently labeled centromere/ kinetochore marker and microtubules. (Figure 1).
