Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
In this experiment we use an artificial bilayer lipid membrane system, featuring the oriented encapsulation of membrane proteins in a functionally active form.
Nickel-nitrilo-tri-acetic acid-functionalized silica nanoparticles of a diameter of around 25 nm are used to attach the proteins via a genetically engineered histidine-tag in a uniform orientation. Subsequently the proteins are reconstituted within a phospholipid bilayer, formed around the particles by in-situ dialysis to form so-called proteo-lipobeads (PLBs). With a final size of about 50 nm, the PLBs can be employed for UV/VIS spectroscopy studies, particularly of multi-redox centre proteins, since effects of light scattering are negligible. As a proof of concept we use cytochrome c oxidase from (CcO) from P. denitrificans with the his-tag genetically engineered to subunit I. (Fig. 1).

Schematics of a proteo-lipobead: Membrane proteins such as CcO (marked in red) are attached to a Ni-NTA-modified central particle (marked in dark grey) via his-tag technology. Phospholipid molecules (marked in light grey) are inserted in-between the proteins to form a freely suspended lipid bilayer membrane.
In this orientation, the P side of CcO is directed to the outside and hence electron transfer can be initiated by illumination of Ruthenium complexes with a flash lamp, using a setup designed to perform time-resolved UV/VIS and alternatively fluorescence spectroscopy (Fig. 2)
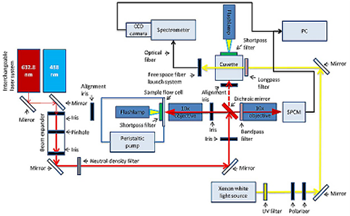
Figure 2: Schematics of the combined timeresolved UV/VIS and fluorescence spectroscopy setup
Figure 2 shows the schematics of the combined time-resolved UV/VIS and fluorescence spectroscopy setup. Basically a dichroic mirror (OMEGA OPTICAL XF2087 660DRLP or XF2010 505DRLP) is used to separate the higher energy probe laser beam, either 632.8 nm from an JDS Uniphase HeNe or a 488 nm Coherent Cube 488-50C diode laser, from the emitted fluorescence light. A 10x objective (Olympus Plan Achromat RMS10X) is used to both focus the laser into the sample as well as collect the fluorescence emission of the material. A second 10x objective of the same type, the coupling objective, is installed behind the dichroic mirror and couples the emitted light into a fibre optic that is connected to the spectrometer (Andor Shamrock SR-303i-B equipped with an Andor CCD detector DU920P-OE). Alternatively, UV/VIS spectra can be collected from a two-way quartz cuvette connected to the xenon flash lamp, which is used to photo-excite the material in the respective sample holder. It is possible to trigger flashes with the function generator (Agilent Technologies 33250a) that can be synchronized with the CCD camera connected to the spectrometer. With this setup it is possible to synchronize light excitation and spectra collection to measure photo induced kinetics.
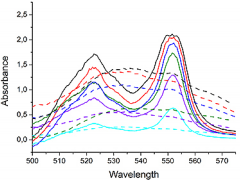
Figure 3: UV/VIS spectra of CcO before (dotted lines) and after (full lines) photoexcitation with the flash lamp
UV/VIS spectra of CcO from Paracoccus denitrificans were measured, initiated by photoexcitation of the Ru complex [(bpy)2Ru(diphen)Ru(bpy)2](PF6)4 (Ru2C). Characteristic bands appear at 523 and 551 nm indicating the reduction of the heme centres of the CcO (Fig. 3).
References
