Resources
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
Expand
Collapse
 Part of the Oxford Instruments Group
Part of the Oxford Instruments Group
In many surgical procedures involving the excision of tumorous tissue, the surgeon is challenged in deciding when the excision is complete whilst at the same time trying to minimise the amount of healthy tissue removed. This is particularly challenging during laparoscopy or keyhole surgery, and even more so if robotic surgery is included, where the view of the tissue and the tactile feedback are restricted, or absent.
Researchers are turning to photonic techniques such as Raman spectroscopy, which can offer powerful detection capability in the field of bio-medical optics that aids in the identification of the chemical constituents of tissues and cells. This in turn can assist the surgeon in making a reliable diagnosis as regards the type of tissue. A group at the University of St Andrews [1,2] have tackled this problem by designing, building and demonstrating a robotic based analytical tool to assist surgeons in such surgical procedures.
Introduction
Many surgical procedures are conducted nowadays with the assistance of robotic systems. Ashok et. al. [1] have developed a Raman probe based system to complement current standard diagnostic techniques such as histological examination. In their design a Raman probe based sensor was integrated into a surgical robot - ARAKNES (Array of Robots Augmenting the KiNematics of Endoluminal Surgery) [3], with the aim of demonstrating a tool for robot-assisted laparoscopic surgery. Laparoscopy is a type of surgical procedure that allows a surgeon to access the inside of the body with keyhole surgery, for example the abdomen, without having to make large incisions in the skin.
Raman spectroscopy can offer complementary information to, and possibly exceeding, purely vision and touch, regarding tissue morphology and chemical composition. This provides a basis for having multimodal information to aid decision making. The use of fibre-based Raman probes has been demonstrated in various studies for in-vivo and ex-vivo tissue analysis [4-6]. Ashok and his co-workers show that such information can improve identification of the boundary or margin between healthy tissue and cancerous tissue during the surgical procedure [1, 2].
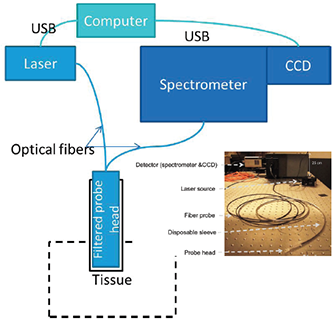
Figure 1: A schematic layout of the optical system showing the main components: a Newton camera, Shamrock SR303i, the diode laser and the Raman probe (Emvision LLC), and probe head with disposable sleeve.
Setup
A schematic of the spectroscopy part of the system is shown in figure 1. The Raman sensor consists of four principal subsystems:
The fibre probe cables were 5 m long to allow for a safe working distance of the spectroscopy system from the operating table, and were terminated by two SMA connectors at the ends connected to the excitation laser and spectrometer. At the other end of the cable was a rigid probe head with length of 50 mm and diameter of 3.8 mm. It contained a low-pass filter at the excitation side and a long-pass filter at the collection side to reduce the background fluorescence from the optical fibre. A GRIN lens mounted at the tip of the probe head set the working distance from the end of the probe head to approximately 1 mm. The resulting size of the excitation beam at the sample was ~ 500 µm in diameter. A number of key features were designed into the probe head to ensure that,
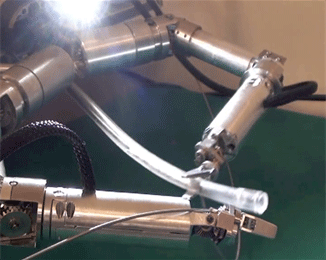
Figure 2: The fiber probe being held and manipulated by the robotic arm of ARAKNES.
A picture of the fibre probe being manipulated by the robot arm is shown in figure 2.
A disposable sterile sleeve, into which the probe was inserted, was used to maintain sterility of the probe during the surgical procedure. A sapphire window was bonded to the end of the sleeve and this allowed optical access to the tissue from the probe head. The probe head was pushed up against the 1 mm thick sapphire window. Since the probe head had a working distance of 1 mm – the sleeve-head assembly was used in contact mode i.e. the sapphire window was brought into contact with the tissue when taking the Raman spectral data, thus minimising any variability in the signal due to different sample-probe distances.
Another very important component of the system was the user-compatible software interface. Clearly the tool has to be user-friendly to surgeons and medical staff, whose expertise is not analytical spectroscopy, but who want to know whether the tissue under test is healthy or cancerous. Ashok et. al. implemented a supervised multivariate classification algorithm to classify the different tissue types. Two key features incorporated within their protocol for analysing the data were:
The interface that performs binary tissue classification was based on a machine learning algorithm, where a training dataset was obtained locally from the same patient going through the operation. The aim of the sensor is to assist the surgeon in identifying the separation line between two different tissue types, when such differentiation is otherwise visually ambiguous. Given the known presence of a tumor prior to excision, it will have two known tissue types (tumor, type-A, and non-tumor, type-B). Hence this gives the opportunity to train the binary classifier for the margin identification task by using sample areas from the two distinct tissue regions from the specific patient concerned. Having trained the system to identify the two different tissue types, it can then be used to identify the unknown tissue type in the regions around the boundary between healthy and cancerous tissue, and thereby allow the surgeon to determine the location of that boundary.
Results and Discussion
In order to test the efficacy of the system, proof-of-principle tests were carried out on excised tissues that were visually similar. Adipose tissues derived from various animals were used in this tissue discrimination analysis. A typical Raman spectrum captured from Bovine adipose tissues is shown in figure 3. With chemometric analysis of the subtle differences between spectral features from different samples the different tissues types can be identified. An accuracy of 95% was obtained in discriminating these tissue types.
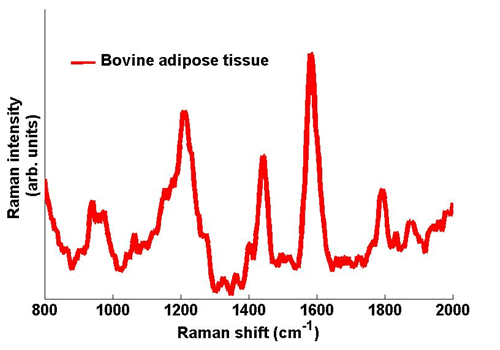
Figure 3: Typical Raman spectra taken from Bovine adipose tissue. The spectrum was captured with an exposure time on the Newton camera of 1 s.
The development of this system represents a significant step forward in the development of robotically assisted surgical tools incorporating on-line photonic based diagnostic techniques. Specifically Raman spectroscopy can provide a wealth of complementary information for tissue discrimination and thereby aid surgeons in making more informed decisions whilst carrying out surgical procedures. The St Andrews group approach has been a modular one which will allow such tools to be used in other platforms. They have developed a protocol which overcomes patient-to-patient variability, and binary discrimination capability which can be applied to a range of different tissue types.
Acknowledgement: Appreciation is gratefully extended to Dr Praveen Ashok for his assistance. Images courtesy of the group at the University of St Andrews.
References:
